Microfluidic flow cells: off-chip pH monitoring for organ-on-a-chip
Author
Camila Betterelli Giuliano, PhD
Publication Date
Keywords
pH Monitoring
Cell culture
Organ-on-a-chip
off-chip monitoring
Microfluidic flow cells

Need advice for your pH monitoring?
Your microfluidic SME partner for Horizon Europe
We take care of microfluidic engineering, work on valorization and optimize the proposal with you
Microfluidic cell culture

Traditionally, the culture of live cells outside the body is performed in monolayers inside static polymer-coated plastic flasks placed in CO2 incubators, which maintain a high humidity, constant temperature (37° C), and CO2 concentration (5%) for pH buffering. The cultures comprise a single cell type to facilitate analysis and avoid cross-contamination [1].
Cell culture provides a platform to study cellular behavior in a systematic way, with proper isolation of variables and straightforward experimental design, allowing investigations that would not be ethical to be conducted in humans while reducing the use of non-analogous animal models.
Moreover, the advent of cell culture has been undeniably responsible for historical achievements in the biomedical area, such as the polio vaccine, the connection between viruses and cancer, the emergence of the genetic engineering field, etc [2].
... A way toward flow cells
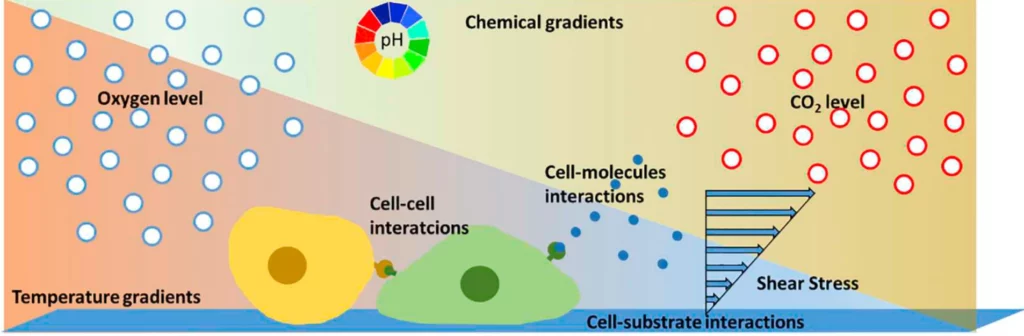
Nevertheless, it is a reductionist approach that fails to represent the intrinsic complexity of cell differentiation, interaction, tissue function, drug response, and the dynamic and intricate microenvironment in vivo biology (Figure 1). These physical and chemical signals are crucial for cellular development and behavior. Their absence leads to poor translation between in vitro and in vivo systems.
Furthermore, traditional cell culture protocols are time-consuming and labor-intensive [3]. These limitations profoundly affect the health and pharmaceutical industries, resulting in inefficient and increasingly costly drug development processes. It is estimated that roughly 40% of new drug candidates that reach clinical trials fail due to unforeseen toxicity effects in humans [4]. Closing the gap between preclinical and clinical data translation calls for better methods to predict human responses.
The advantages of microfluidic cell culture, which can easily mimic the cellular microenvironment and provide a high level of control in addition to automated monitoring and analysis, led to the development of the “organ-on-a-chip” (OoC) field in the 2010s, currently one of the most relevant areas of research in microfluidics [6].
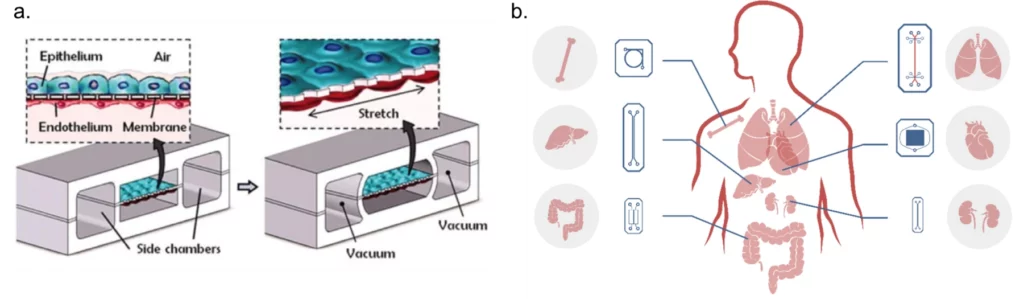
OoCs are specialized in vitro cell culture models based on microfluidic devices that are structured around three main pillars: reproducing the mechanical and biochemical stimulus of the cellular microenvironment, simulating the 3D microarchitecture of tissues with multiple cell types placed in specific communicating compartments, and replicating functional tissue-tissue interfaces via cell-cell interaction (Figure 2a).
The main goal is to reproduce complex in vivo biology to better understand cellular behavior and metabolism, disease phenotypes, and drug response without the need for insufficient in vitro models or imprecise animal models [7].
Several organ-on-a-chip devices have been successfully developed, namely, liver [8], lung [7,9], kidney [10], heart [11,12], intestine [13], bone [14], bone marrow [15], nerve [16], blood vessels [17, 18], and blood-brain barrier [19] (Figure 2b).
They have demonstrated drug responses closer to human physiology than standard in vitro models, with the enhanced complexity leading to better cell differentiation and drug transport [20]. As OoC models mature, the interconnection of several OoCs in a “human-on-a-chip” approach becomes feasible and has been investigated by several groups in different configurations [21-24].
An emerging area that has the potential to profoundly change modern medicine is the development of OoC with induced pluripotent stem cells derived from patients. This could potentially be a paradigm shift in personalized medicine and the development of patient-specific treatments.
The increase in complexity of cell cultures enabled by microfluidics opened the possibility of more in-depth spatiotemporal analysis of cellular development and behavior, further increasing the importance of tools such as live-cell imaging [25], metabolic detection and analysis [26] and microenvironmental monitoring of parameters such as pH [27].
Consequently, the long-term culturing of cells in OoCs moved from the CO2 incubator to the microscope stage. As the complexity of the systems increased and the access to more detailed information deepened, enhanced resolution and real-time monitoring of the systems became a clear demand [26, 28].
Thus, a growing need to monitor temperature, pH, and dissolved O2 more closely, combined with increased interest in understanding the in situ metabolic activity, led to many on-chip and off-chip sensing solutions.
The importance of pH monitoring
As previously mentioned, the gap between preclinical and clinical data causes enormous losses for the health sector, besides delaying patient treatment access. One of the contributing factors to this global issue has been attributed to the variables of microenvironment conditions [29].
Cell lines require a stable pH to grow, as acidic pH can irreversibly inhibit enzyme activities and DNA, RNA, and protein synthesis, thus compromising cell viability [30, 31]. pH is defined as the concentration of free H+ ions in a solution, also called the activity of protons, and can be calculated by the Henderson-Hasselbalch equation:
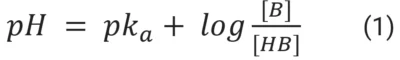
in which pka is the acid dissociation constant, [HB] is the concentration of a compound’s protonated form, and [B] is the concentration of a compound’s unprotonated form.
Buffering systems can maintain the pH constant due to a dynamic equilibrium. The equilibrium between the protonated and unprotonated forms of the buffer can move towards reacting to new free H+ or releasing H+ ions to compensate for added or removed ones from the surrounding solution.
Both mechanisms keep the previous free H+ concentration constant to a certain extent. If the addition or removal of free H+ goes beyond a threshold that the buffer can absorb, the pH will change, indicating that the solution’s buffering capacity has been surpassed [32].
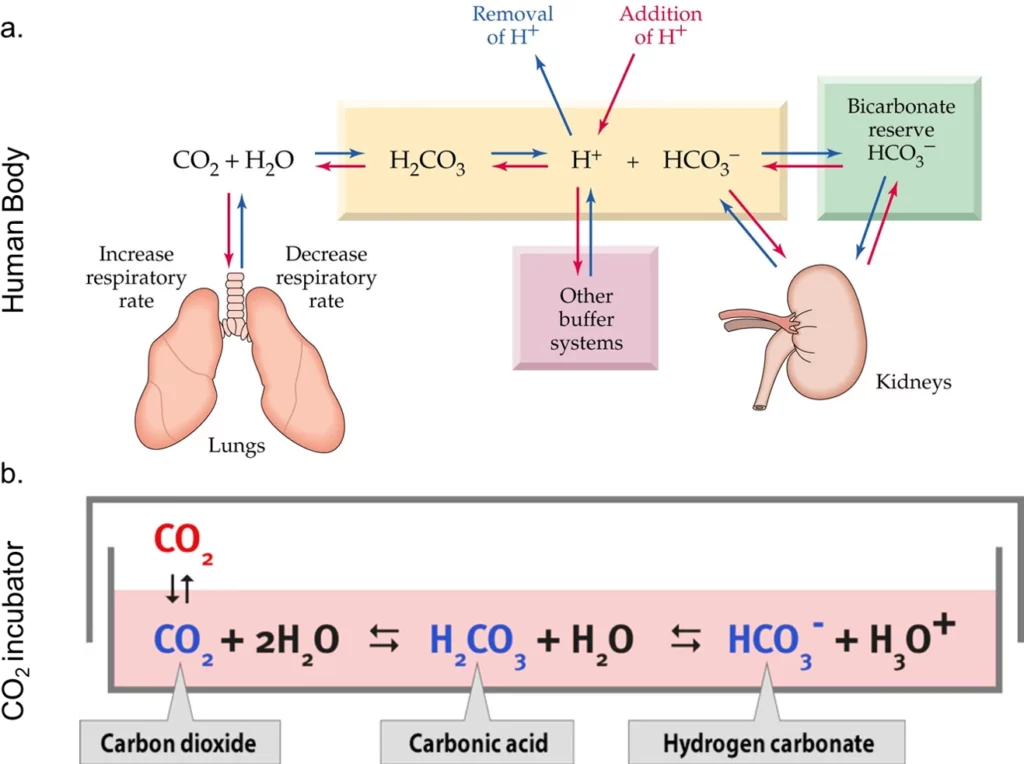
That is why complex organisms, such as mammalians, have developed dynamic buffering systems that adjust the concentration of the buffer according to the system’s needs. The physiological buffering system is based on CO2/HCO3– equilibrium (bicarbonate buffering system, pka= 6.15).
The lungs, through the gas exchange of CO2, and the kidneys, through ion transport proteins, are responsible for keeping the ratio of the protonated to unprotonated species of this buffering system in homeostasis and the organism’s pH within the physiological range (Figure 3a).
Cell metabolism is bound to acidify the pH of the microenvironment due to the production and release of lactate, which reacts with water to form acid lactic. For this reason, cell culture media usually contains a buffering system to keep pH under physiological conditions [33].
Besides the CO2/HCO3– buffer, media can be buffered by non-volatile buffers (NVB), such as HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (pKa = 7.3; 37 °C), PIPES (piperazine-N,N′-bis(2-ethanesulphonic acid); pKa = 6.7) and MES (2-(N-morpholino)-ethanesulfonic acid; pKa = 6.0) [34]. However, NVBs in cell culture media are out of the scope of this review; detailed work on the topic can be found in Michl et al., 2019 [35].
The bicarbonate buffering system is replicated in cell biology labs with a CO2 incubator. This system works through the equilibrium of CO2 from the CO2-rich atmosphere of the incubator, which dissolves in the media and reacts with H2O, forming carbonic acid and the NaHCO3 present in the media (Figure 3b).
Cell culture media come with different concentrations of NaHCO3 and require different percentages of CO2. For example, DMEM comes with 44 mM of NaHCO3, which requires approximately 10% of CO2 in the atmosphere to maintain a pH close to 7.4. CO2 incubators are conventionally set to 5% CO2, keeping the DMEM’s pH around 7.6-7.8. The production of lactic acid and CO2 by healthy cells and the buffering capacity of serum, often added to DMEM, partially offset this difference, allowing the growth of cells with DMEM using conventional CO2 concentrations [35].
However, the implications of this adjustment are not well understood. Cells’ intracellular pH is in close equilibrium with the microenvironment’s extracellular pH due to the transmembrane ion transporter proteins. Michl et al. [35] studied the effect of extracellular pH change on intracellular pH and concluded that cells re-balance the internal pH based on the external one. Since most H+ targets are intracellular, this change can unpredictably affect intracellular mechanisms and metabolic pathways.
Monitoring pH becomes even more critical when the culturing system is downscaled to micron-range in OoCs, substantially reducing the volumes. Slight variations in pH, e.g., 7.4±0.3, are tolerated in traditional cell culture; however, the more accentuated diffusional processes of microfluidic cell culture result in a more pronounced effect on cell viability, prompting closer monitoring of crucial environmental parameters [31].
Monitoring cell culture environment with flow cells
The measurement of metabolic activity in microfluidic cell culture is usually done with on-chip solutions. For example, Weltin et al. [36] developed a microphysiometer system in a microfluidic chip with dissolved O2 and pH electrochemical sensors inside the culturing chamber.
The authors successfully cultured brain cancer cells with online and continuous measurement of the cells’ metabolism. However, these solutions prove too complex and costly [37] when the goal is to monitor the cell culture environment, which does not require the same level of detailed information. For that, microfluidic off-chip solutions, such as microfluidic flow cells, are more apt.
Microfluidic flow-through cells can be developed in-house [38-40], usually 3-D printed [41], or commercially available [42-46]. The format can vary according to the application, such as T-junctions for HPLC applications or other geometrical structures fitted to house the sensor (Figure 4). The main goal is to align the sensors with the microfluidic system to allow continuous monitoring.
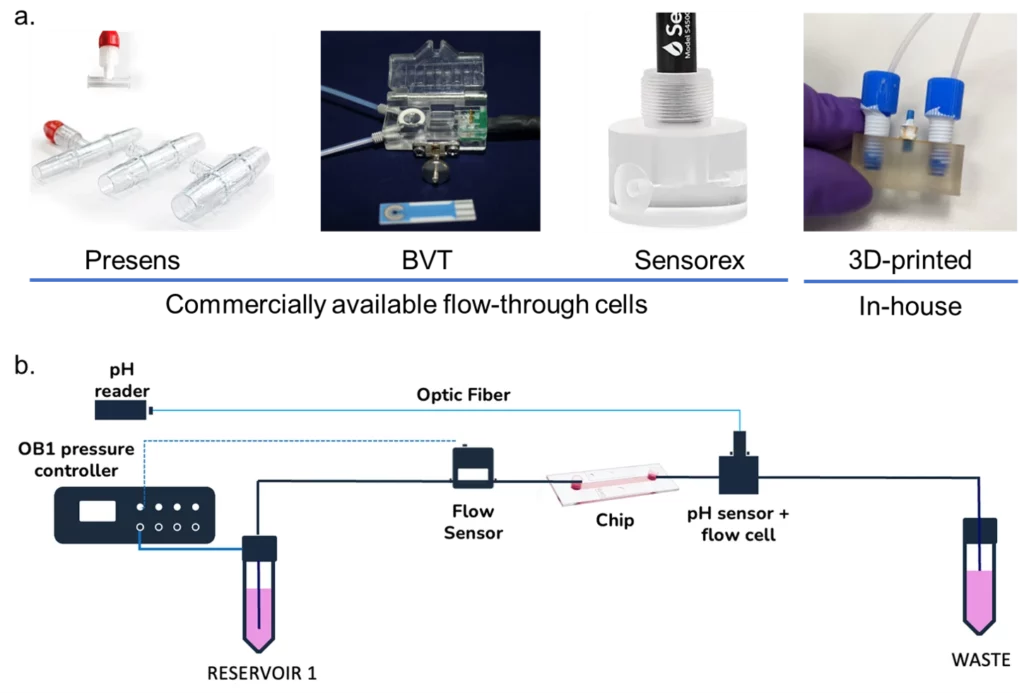
A good example is the work of Zhang et al. [47], who developed a “physical sensing unit” composed of pH, O2, and temperature sensors in a flow-through cell developed in-house to monitor the microenvironment of a complex organ-on-a-chip system.
The system was designed as a modular platform to control two organ-on-a-chip connected to an automated flow control breadboard. Interestingly, the biomarker sensing unit, i.e., the sensors to measure the metabolism, was also off-chip. The platform was validated with a drug screening assay using liver and heart organ-on-a-chip, exposed to acetaminophen for five days and doxorubicin for 24 hours, where cytotoxicity biomarkers were detected and quantified.
Farooqi et al. [26] 3-D printed a pH-sensing flow-through cell to house an optical pH sensor built with commercially available parts that measured pH based on the color of the red phenol in the media. The goal was to monitor a live-on-chip system also intended to investigate the drug toxicity of doxorubicin. The researchers detected a decrease in pH when cells were exposed to high drug concentrations compared to the control. The same group performed similar work with a lung-on-chip system [48], attesting to the robustness of the developed sensing unit.
Ali et al. [49] similarly monitored fibroblasts’ pH and O2 for three days. Wu et al. [50] designed a high-throughput pH sensing unit consisting of parallelized microfluidic channels with a sensing chamber connected to optical pH sensors using the medium’s phenol red. They performed simulations to define the best shape of the sensing chamber, resulting in an oval shape.
The thickness of the polydimethylsiloxane (PDMS) layer has also been decreased to minimize the loss of light transmission. Optical sensors seem the most common choice for microfluidics flow-through cells due to their independence of reference electrodes, electrical connections, and flow rates. They are also not prone to biofouling, corrosion, and interference from electrochemical signals from molecules in the medium [49].
Conclusion on off-chip flow cell for monitoring
The important growth of organ-on-a-chip technologies increased the need to monitor what was happening inside the chips more closely. Several on-chip and off-chip solutions have been designed, and microfluidic flow cells seem to be the way forward for microenvironment pH monitoring.
On-chip sensors are suited for detecting metabolic activity that can benefit from high spatiotemporal resolution. However, these solutions are too costly and complex for microenvironment monitoring, which does not require the same level of precision. For microenvironment monitoring, microfluidic flow cells with embedded optical sensors placed in the microfluidics circuit for real-time and continuous monitoring have been the most common choice.
Review done thanks to the support of the Protomet H2020-MSCA-ITN-2018-Action “Innovative Training Networks”, Grant agreement number: 813873
Author: Camila Betterelli Giuliano, PhD
Contact:
Partnership[at]microfluidic.fr


References
- Coluccio, M. L. et al. Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 208, 14–28 (2019).
- Lyapun, I. N., Andryukov, B. G. & Bynina, M. P. HeLa Cell Culture: Immortal Heritage of Henrietta Lacks. Mol. Genet. Microbiol. Virol. 34, 195–200 (2019).
- Mehling, M. & Tay, S. Microfluidic cell culture. Curr. Opin. Biotechnol. 25, 95–102 (2014).
- Van Norman, G. A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach? JACC Basic to Transl. Sci. 4, 845–854 (2019).
- Berthier, E., Surfus, J., Verbsky, J., Huttenlocher, A. & Beebe, D. An arrayed high-content chemotaxis assay for patient diagnosis. Integr. Biol. 2, 630–638 (2010).
- Convery, N. & Gadegaard, N. 30 years of microfluidics. Micro Nano Eng. 2, 76–91 (2019).
- Huh, D. et al. Reconstituting organ-level lung functions on a chip. Science (80-. ). 328, 1662–1668 (2010).
- Novik, E., Maguire, T. J., Chao, P., Cheng, K. C. & Yarmush, M. L. A microfluidic hepatic coculture platform for cell-based drug metabolism studies. Biochem. Pharmacol. 79, 1036–1044 (2010).
- Stucki, A. O. et al. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip 15, 1302–1310 (2015).
- Jang, K. J. et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 5, 1119–1129 (2013).
- Grosberg, A., Alford, P. W., McCain, M. L. & Parker, K. K. Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip. Lab Chip 11, 4165–4173 (2011).
- Kobuszewska, A., Jastrzębska, E., Żukowski, K. & Brzózka, Z. Simulation of hypoxia of myocardial cells in microfluidic systems. Sci. Reports 2020 101 10, 1–11 (2020).
- Kim, H. J. & Ingber, D. E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 5, 1130–1140 (2013).
- Mansoorifar, A., Gordon, R., Bergan, R. C. & Bertassoni, L. E. Bone-on-a-Chip: Microfluidic Technologies and Microphysiologic Models of Bone Tissue. Adv. Funct. Mater. 31, 2006796 (2021).
- Torisawa, Y. S. et al. Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat. Methods 2014 116 11, 663–669 (2014).
- Shi, M. et al. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab Chip 13, 3008–3021 (2013).
- Van Der Meer, A. D., Orlova, V. V., Ten Dijke, P., Van Den Berg, A. & Mummery, C. L. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip 13, 3562–3568 (2013).
- Yu, F., Selva Kumar, N. D., Choudhury, D., Foo, L. C. & Ng, S. H. Microfluidic platforms for modeling biological barriers in the circulatory system. Drug Discov. Today 23, 815–829 (2018).
- Booth, R. & Kim, H. Characterization of a microfluidic in vitro model of the blood-brain barrier (μBBB). Lab Chip 12, 1784–1792 (2012).
- Bhatia, S. N. & Ingber, D. E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014 328 32, 760–772 (2014).
- Novak, R. et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat. Biomed. Eng. 4, 407–420 (2020).
- Zhang, C., Zhao, Z., Abdul Rahim, N. A., Van Noort, D. & Yu, H. Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments. Lab Chip 9, 3185–3192 (2009).
- Luni, C., Serena, E. & Elvassore, N. Human-on-chip for therapy development and fundamental science. Curr. Opin. Biotechnol. 25, 45–50 (2014).
- Renggli, K., Rousset, N., Lohasz, C., Nguyen, O. T. P. & Hierlemann, A. Integrated Microphysiological Systems: Transferable Organ Models and Recirculating Flow. Adv. Biosyst. 3, 1900018 (2019).
- Schneckenburger, H. & Richter, V. Challenges in 3d live cell imaging. Photonics 8, (2021).
- Kim, B. S. et al. Real-time physiological sensor-based liver-on-chip device for monitoring drug toxicity. J. Micromechanics Microengineering 30, 115013 (2020).
- Kilic, T., Navaee, F., Stradolini, F., Renaud, P. & Carrara, S. Organs-on-chip monitoring: sensors and other strategies. Microphysiological Syst. 1, 1–1 (2018).
- Santbergen, M. J. C., van der Zande, M., Bouwmeester, H. & Nielen, M. W. F. Online and in situ analysis of organs-on-a-chip. TrAC – Trends Anal. Chem. 115, 138–146 (2019).
- Begley, C. G. & Ellis, L. M. Drug development: Raise standards for preclinical cancer research. Nature 483, 531–533 (2012).
- Morita, T., Nagaki, T., Fukuda, I. & Okumura, K. Clastogenicity of low pH to various cultured mammalian cells. Mutat. Res. Mol. Mech. Mutagen. 268, 297–305 (1992).
- Lu, C. & Verbridge, S. S. Microfluidic methods for molecular biology. Microfluid. Methods Mol. Biol. 1–376 (2016). doi:10.1007/978-3-319-30019-1
- Scorpio, R. Fundamentals of Acids, Bases, Buffers & Their Application to Biochemical Systems. (Kendall Hunt Pub Co, 2000).
- Salis, A. & Monduzzi, M. Not only pH. Specific buffer effects in biological systems. Curr. Opin. Colloid Interface Sci. 23, 1–9 (2016).
- Eagle, H. Buffer combinations for mammalian cell culture. Science (80-. ). 174, 500–503 (1971).
- Michl, J., Park, K. C. & Swietach, P. Evidence-based guidelines for controlling pH in mammalian live-cell culture systems. Commun. Biol. 2019 21 2, 1–12 (2019).
- Weltin, A. et al. Cell culture monitoring for drug screening and cancer research: a transparent, microfluidic, multi-sensor microsystem. Lab Chip 14, 138–146 (2013).
- Fuchs, S. et al. In-line analysis of organ-on-chip systems with sensors: Integration, fabrication, challenges, and potential. ACS Biomater. Sci. Eng. 7, 2926–2948 (2021).
- Pousti, M., Zarabadi, M. P., Abbaszadeh Amirdehi, M., Paquet-Mercier, F. & Greener, J. Microfluidic bioanalytical flow cells for biofilm studies: a review. Analyst 144, 68–86 (2018).
- Chauvet, A., Tibiletti, T., Caffarri, S. & Chergui, M. A microfluidic flow-cell for the study of the ultrafast dynamics of biological systems. Rev. Sci. Instrum. 85, 103118 (2014).
- Akin, M. et al. A new set up for multi-analyte sensing: At-line bio-process monitoring. Biosens. Bioelectron. 26, 4532–4537 (2011).
- Capel, A. J., Rimington, R. P., Lewis, M. P. & Christie, S. D. R. 3D printing for chemical, pharmaceutical and biological applications. Nat. Rev. Chem. 2, 422–436 (2018).
- Strebl, M. G., Bruns, M. P., Schulze, G. & Virtanen, S. Respirometric In Situ Methods for Real-Time Monitoring of Corrosion Rates: Part II. Immersion. J. Electrochem. Soc. 168, 011502 (2021).
- Ahmerkamp, S. et al. The effect of sediment grain properties and porewater flow on microbial abundance and respiration in permeable sediments. Sci. Reports 2020 101 10, 1–12 (2020).
- Higuera, G. A. et al. Supporting data of spatiotemporal proliferation of human stromal cells adjusts to nutrient availability and leads to stanniocalcin-1 expression in vitro and in vivo. Data Br. 5, 84–94 (2015).
- Illner, S., Hofmann, C., Löb, P. & Kragl, U. A Falling-Film Microreactor for Enzymatic Oxidation of Glucose. ChemCatChem 6, 1748–1754 (2014).
- Krejčí, J. et al. The measurement of small flow. Sensors Actuators, A Phys. 266, 308–313 (2017).
- Zhang, Y. S. et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U. S. A. 114, E2293–E2302 (2017).
- Khalid, M. A. U. et al. A lung cancer-on-chip platform with integrated biosensors for physiological monitoring and toxicity assessment. Biochem. Eng. J. 155, 107469 (2020).
- Ali, S., Shaegh, M., De Ferrari, F. & Zhang, Y. S. A microfluidic optical platform for real-time monitoring of pH and oxygen in microfluidic bioreactors and organ-on-chip devices. Biomicrofluidics 10, 044111 (2016).
- Wu, M. H., Lin, J. L., Wang, J., Cui, Z. & Cui, Z. Development of high throughput optical sensor array for on-line pH monitoring in micro-scale cell culture environment. Biomed. Microdevices 11, 265–273 (2009).
Check the other Reviews
FAQ - Microfluidic flow cells: off-chip pH monitoring for organ-on-a-chip
What is the importance of pH measurement in organ-on-chip systems?
Cell viability is sensitive to constant pH (typically 7.4 ± 0.3). Alkaline environments irreversibly suppress the activities of enzymes and DNA/RNA/protein synthesis. In microfluidic devices with small volumes, drastic pH changes have strong effects because diffusional processes are exaggerated by the small volumes; therefore, constant monitoring is critical to maintain physiological state and ensure valid experimental outcomes.
How does the bicarbonate buffering system occur in cell culture?
Physiological buffering is mimicked by the system using the CO2/HCO3 equilibrium. CO2 incubators: CO2 present in the atmosphere dissolves within the media, and reacts with H 2 O to produce carbonic acid and balances with NAHCO3 in the press. The 5% CO2 is used in traditional culture, but some media, such as the DMEM, need approximately 10% CO2 to reach optimal pH. Lactic acid and CO2 are naturally produced in cells and are taken up by the buffer system to maintain homeostasis.
What are the microfluidic flow cells, and how do they operate?
Microfluidic flow cells are equipment that align microfluidic circuits with sensors, allowing them to be observed in real time. They are custom components that can be 3D-printed or purchased off the shelf, with geometries ranging from simple T-junctions to tailored sensor enclosures. The cell is subjected to media that passes through embedded sensors to provide inline readings without interfering with cell culture.
What benefits do optical sensors have in flow cell pH monitoring?
pH is detected using optical sensors that change the color of the medium based on phenol red. The main benefits are that it is independent of reference electrodes, requires no electrical connection, is not affected by biofouling or corrosion, does not interfere with electrochemical signals in the media, and is independent of flow rate. This enables them to be stronger and less expensive than electrochemical methods for microenvironment monitoring.
Why do off-chip flow cells tend to be a better choice as compared to on-chip sensors to monitor pH?
On-chip sensors offer high spatiotemporal resolution, which is useful for detailed metabolic studies, but are somewhat complex and expensive. Off-chip flow cells are sufficiently precise for microenvironment monitoring (though not as accurate as microfluidics) at much lower cost and complexity, making them practical for experiments involving routine organ-on-chip approaches.
What is the role of MIC in the integration of pH monitoring of organ-on-chip platforms?
MIC offers full integration of the system, including: design of custom flow cells via 3D printing, a combination of optical and electrochemical pH sensors, fine flow control systems with minimal pulsation to enable stable measurements, and platforms for real-time data acquisition and cloud integration.
What are the fabrication methods that MIC can propose to create flow-through cells?
MIC has many paths to fabricating: high-resolution SLA/DLP 3D printing of complex structures to make prototypes; PDMS soft lithography of transparent structures; biocompatible designs; thermoplastic molding to produce structures in large quantities; and micro-CNC/laser machining of precision structures.
Is it possible to have multiple non-pH sensors, as well as pH, in flow cell systems with MIC?
Yes, MIC creates multi-parameter sensing platforms that combine: temperature, pressure, O2 sensors, optical detection of fluorescence/absorbance, electrochemical impedance sensors, metabolite detection, and multi-channel calibration signal processing.
What is the MIC solution for fully covering the chip-to-data analysis?
The solutions offered by MIC include organ-on-chip design with a physiological microenvironment, fluidic control with automated protocols, embedded electronics and firmware for sensor integration, software stacks with GUI and LIMS integration, and data engineering to standardize and train AI.
What are the quality control measures that MIC uses on pH monitoring systems?
MIC uses extensive validation: sensor correctness and drift testing, stress testing under environmental conditions, multi-parameter testing, and metrology: experimental design and uncertainty analysis.
Will MIC help with scholarly studies as well as with the business development of monitoring systems?
Absolutely. MIC also closes the research-to-market gap with bespoke sensor development tailored to research needs, Design for Manufacturing (DfM) to production at scale, regulatory compliance for medical devices, and consortium management expertise for EU-funded projects.
What is the integration of AI in MIC systems in pH and organ-on-chip systems?
MIC uses AI throughout the development chain: adaptive control algorithms based on real-time pH sensor feedback to control flow conditions, automated experiment design based on optimization of culture parameters, computer vision based on correlations between pH changes and cellular morphology, and rare-event detectors based on the identification of pathological responses.
So, in cases where multi-organ systems require coordinated monitoring, how is MIC used?
MIC architects are complex by: system architecture capabilities spanning from application requirements to full integration; multi-channel flow control of parallelized organs; modular sensing units scalable across various organ-on-chip configurations; and software that synchronizes data across multiple organs.
What is the difference between the approach of MIC to organ-on-chip monitoring solutions?
MIC has a high level of integration, not as isolated components: physics-based models, experimental validation, expertise in fabrication, comprehensive instrumentation and sensing, regulatory compliance, built-in from the first design, and systems thinking that treats chips, instruments, software, and data as integrated platforms.