Spatial Transcriptomics with Automated Fluidic Control
Plug-and-play fluidic automation for merfish/seqfish/seqfish+
Instruments for spatial transcriptomics
The perfect pack for your fluorescence in situ hybridization setup
Intergration and automation
Perform automated personalized sequences and microscope synchronisation through TTL triggers and SDK
Easy sequential injection
Quickly swap between solutions with only one flow controller

Need a microfluidic SME partner for your Horizon Europe project?
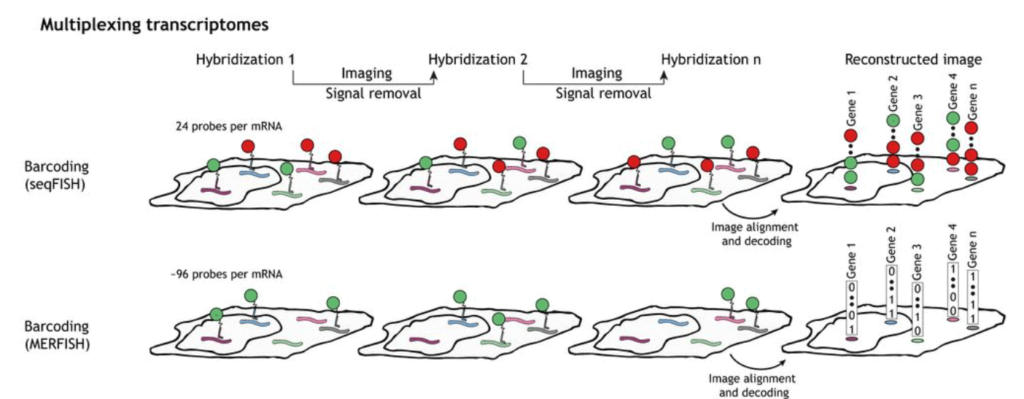
Sequential hybridization figures by Lubeck et al. (2014) and Shah et al. (2016)
Fluidic spatial transcriptomics
Quickly implement your MERFISH/seqFISH/seqFISH+ experiments with this all-included, user-friendly, customizable, and automatable spatial transcriptomics pack.
A state-of-the-art flow controller, coupled with other instruments, like selecting valves, is ideally suited to precisely and automatically dispense tens of dyes, imaging, and washing solutions. This pack is more cost-efficient, practical, and easy-to-use than syringe pumps. This platform is also highly flexible and versatile, as it can quickly adapt to the FISH experiment’s required solutions.
This pack aims to allow you to perform multiplexed fluorescence in situ hybridization at a microfluidic scale, significantly reducing the cost of each experiment by reducing the amount of solution. It also allows the system to be automated and improves the reproducibility of results.
This easy-to-use microfluidic spatial transcriptomics for MERFISH/seqFISH pack contains several critical functions, including a precise and perfectly controlled flow rate for precise small dispensed volume, a fast and easy sequential injection between different solutions, and flexible software for sequence setting and automation with the built-in automation sequencer.
The sequence scheduler transforms the setup into an all-in-one automatized platform that can easily flow many different solutions for your spatial transcriptomics and can further be synchronized with other scientific equipment, such as a fluorescence microscope.
Contact our experts for help integrating your experiment using TTL triggers or direct software integration via SDK.
A microfluidic chip is used to perform spatial transcriptomics. Chips are transparent, available in several materials and shapes, with different numbers of inlets/outlets, and compatible with varying FISH methods. We can provide different chips with this pack with specific channel heights, widths, lengths, and materials. Spatial transcriptomics analysis can be expanded even further by using several microfluidic chips in parallel or one microfluidic chip with several channels.
Our all-in-one pack guarantees good compatibility between the different instruments, allows you to start your experiment right away, is piloted by a single software, and can be used for other applications. We also provide continuous and full customer support to fulfill your experiment goals.
Spatial transcriptomics pack setup
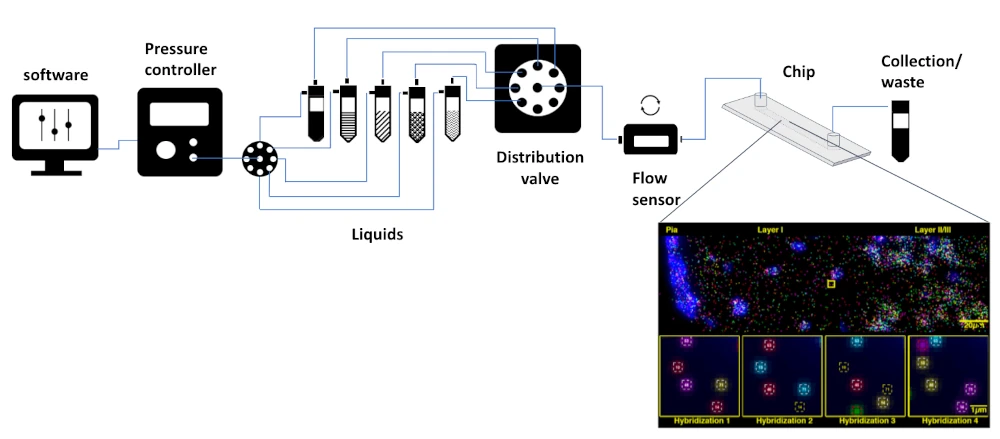
A typical pack contains:
Flow sensor (Galileo, MIC)
Software (Galileo user interface)
Pressure controller
Distribution valve
Tubings and luers
Several Eppendorfs or Falcon reservoirs
Microfluidic chips
A user guide
Spatial transcriptomics principle
The field of research that studies spatially resolved transcriptomes has recently been gaining interest to expand our knowledge of complex multicellular biological systems and their interactions [2]. It has also been chosen by Nature Methods as the “Method of the Year” for 2020 [3].
Cells can sense neighboring cells and their local environment through molecular and cellular mechanisms; thus, it is crucial to study complex biological phenomena to measure the phenotypic and genomic states of single cells in their respective spatial positions [4].
SeqFISH and MERFISH are in situ hybridization encoding methods that use probe detection for single-cell spatial transcriptomics [5]. They are an improved version of the smFISH technique used to study gene expression variations and their consequences [6]. Other methods have also been developed at an increasing rate in the last few years to obtain spatially resolved transcriptomes [7].
To perform seqFISH, a first in situ hybridization is done with one set of fluorescent FISH probes and a labeled dye. DNase is then used to remove the fluorophores. The mRNA is hybridized with the identical FISH probes but labeled with another dye. With several dyes and rounds of hybridization, it is possible to barcode many genes in single cells [8].
SeqFISH has been improved to seqFISH+, which can be used for cell spatial and biological process studies by combining seqFISH with a confocal microscope. This was used to perform super-resolution imaging and multiplexing of 10,000 genes in a single cell [9].
Multiplexed Error Robust Fluorescence In Situ Hybridization (MERFISH) is an improvement to the single-molecule Fluorescence In Situ Hybridization (smFISH) method by massively parallelizing and identifying hundreds to thousands of RNA species simultaneously with spatial information.
This method is also error-robust, thanks to using unassigned binary barcodes that can detect and correct errors. This is the main difference between it and seqFISH, coded in a color sequence [10].
Using lab-on-chip technologies and microfluidic platforms provides several improvements to seqFISH and MERFISH. Mainly to reduce the cost, which can be pretty high for these methods, the experiment time, and to automate the process [11].
References
Shah, Sheel & Lubeck, Eric & Zhou, Wen & Cai, Long. (2016). In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 92. 342-357.
Asp M., Bergenstrahle J., Lundeberg J., Spatially Resolved Transcriptomes—Next Generation Tools for Tissue Exploration, BioEssays, 2020, 42 (10)
Marx, V. Method of the Year: spatially resolved transcriptomics. Nat Methods 18, 9–14 (2021).
Mayr U., Serra D., Liberali P. Exploring single cells in space and time during tissue development, homeostasis and regeneration. Development, 2019, 146(12),
Zhou Y, Jia E, Pan M, Zhao X, Ge Q. Encoding Method of Single-cell Spatial Transcriptomics Sequencing. Int J Biol Sci 2020; 16(14):2663-2674.
Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226.
Asp, M., Bergenstråhle, J., Lundeberg, J., Spatially Resolved Transcriptomes—Next Generation Tools for Tissue Exploration. BioEssays 2020, 42, 1900221.
Lubeck, E., Coskun, A., Zhiyentayev, T. et al. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods 11, 360–361 (2014).
Eng, CH.L., Lawson, M., Zhu, Q. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+. Nature 568, 235–239 (2019).
Moffitt, J R, and X Zhuang. “RNA Imaging with Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH).” Methods in enzymology vol. 572 (2016): 1-49.
Rodriguez-Mateos, P., Azevedo, N.F., Almeida, C. et al. FISH and chips: a review of microfluidic platforms for FISH analysis. Med Microbiol Immunol 209, 373–391 (2020).
Why use microfluidics for fluorecence in situ hybridization?
Using microfluidics is the most efficient method to perform MERFISH (Multiplexed Error-Robust Fluorescence In Situ Hybridization) or seqFISH (sequential Fluorescence In Situ Hybridization) and observe multiple genes and their spatial configuration. It permits experimentation with a drastically lower amount of expensive dye and buffer solutions and is compatible with biological applications and microscope observations.
As mentioned above, an automatized sequence can be implemented to inject the solutions into the cell. Furthermore, several different chips can be connected to the system to observe different samples in parallel easily.
This pack can also be combined with other microfluidic steps before this fluorescent in situ hybridization setup; for single-cell isolation, for example, you can use microfluidic single-cell encapsulation [1].
Microfluidics can also be used for the MA-FISH method, which uses oscillatory flows of diluted probe solutions or performs barcoding (DBiT-seq).
The Microfluidics Innovation Center has developed expertise in microfluidics over more than ten years. It can provide its state-of-the-art expertise in biology and engineering, making it the perfect partner for your transition to microfluidics.
References
1. Mayr U., Serra D., Liberali P. Exploring single cells in space and time during tissue development, homeostasis and regeneration. Development, 2019, 146(12),
Customize your fluidic control for spatial transcriptomics pack
This pack is highly customizable to fit your experiment specification and spatial transcriptomics methods (MERFISH, seqFISH,…).
If you’re unsure about the instrument choices and settings best suited for your application, contact one of our experts!