Droplets encapsulation for biological applications: a review
Author
Camila Betterelli Giuliano, PhD
Publication Date
July 23, 2020
Keywords
Droplet encapsulation
microreactors
Artificial cells
High-throughput screening
Drug delivery

Need advice for your droplet encapsulation?
Your microfluidic SME partner for Horizon Europe
We take care of microfluidic engineering, work on valorization and optimize the proposal with you
Droplet encapsulation creates microreactors to explore heterogeneity in biology
As biologists dive deep into the intricacies of cell and molecular biology, it becomes ever more apparent that many heterogeneities were being overlooked in traditional bulk analysis and experiments.
Exploring these subtleties without losing the speed and accuracy of conventional protocols is becoming a perfect application for microfluidics, especially encapsulation in droplets [1].
Droplet-based microfluidics are micrometer-sized droplet emulsions created in a microfluidic device. Using droplets in research is no novelty, but miniaturizing them to the microscale offers excellent advantages due to their larger surface-to-volume ratio.
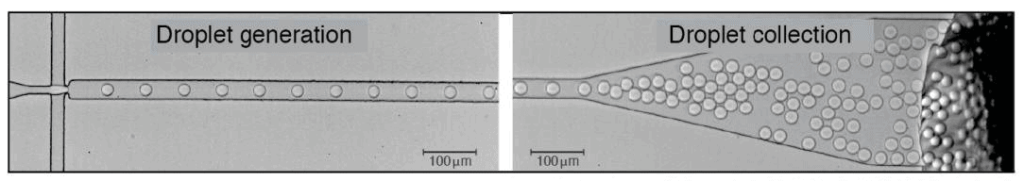
This review will focus on the droplet encapsulation of samples and the potential applications of these microreactors in biology. You can check out our review for more information on droplet-based microfluidics.
Most used droplet encapsulation techniques
Droplet encapsulation techniques vary according to several factors, for example, the type and number of samples needed inside a single droplet and the desired order and time of mixing for chemical reactions. The most common form of encapsulation is to dilute the sample in the solution that will become the dispersed phase, in other words, the solution that will be turned into droplets [4].
Encapsulation during droplet formation
In single-cell droplet encapsulation, diluting the cells into the droplets’ dispersed phase before droplet formation is the most used method. The basic principle is to cut the cell suspension enough so that two cells will unlikely be inside the same droplet.
This approach is simple but wasteful, as it generates a lot of empty droplets since the probability of having one cell inside one droplet is relatively low: 36,8%. If there is a need to encapsulate two distinct cell types in the same droplet, or a cell and a bead, the probability of having one droplet with only one of each entity falls to 13,5%. It generates even more waste due to the higher dilution of the dispersed phase [1].
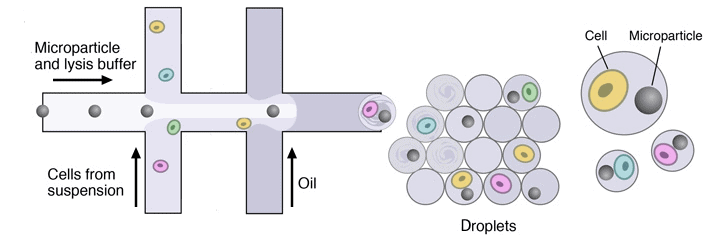
As mentioned in the two-cell example above, it is possible to have two different sample inlets as dispersed phases in a co-flow design, which will mix once they become a droplet [5].
It is also possible to merge two droplets that contain different reagents through electro-coalescence [6], which destabilizes the surface tension of surfactants so the two droplets can coalesce when in contact, or acoustic tweezer, which holds a droplet in place so flowing droplets can collide and merge into it [7].
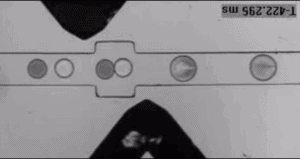
Encapsulation after droplet formation
Lastly, it is possible to inject reagents into existing droplets, giving more manipulation flexibility after droplet formation. There are two possibilities: plug-based and picoinjection. The plug-based microfluidic design uses a channel perpendicular to the flow of droplets to inject the new reagent via an injection droplet into the already-formed droplets [8]. And the picoinjection follows a similar procedure but uses an actuated electric field to have a sub-picoliter control of the injection [9].
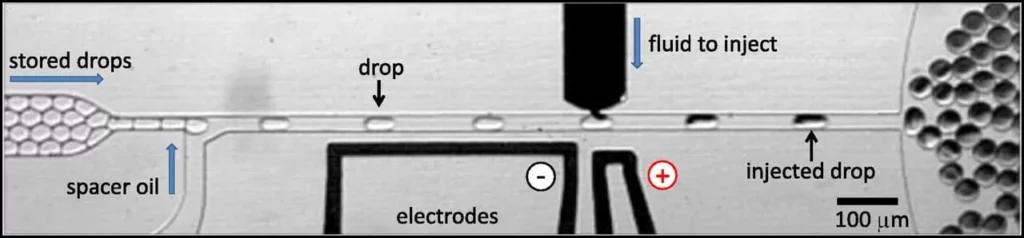
Droplet encapsulation as a tool for biological applications
High-throughput screening based on droplet encapsulation
High-throughput screening is a method of analyzing large libraries of compounds in a short time frame, for example, between 104 and 105 daily tests [11]. It has been widely used in drug discovery, toxicity, and antibody affinity assays with robotics and well plates.
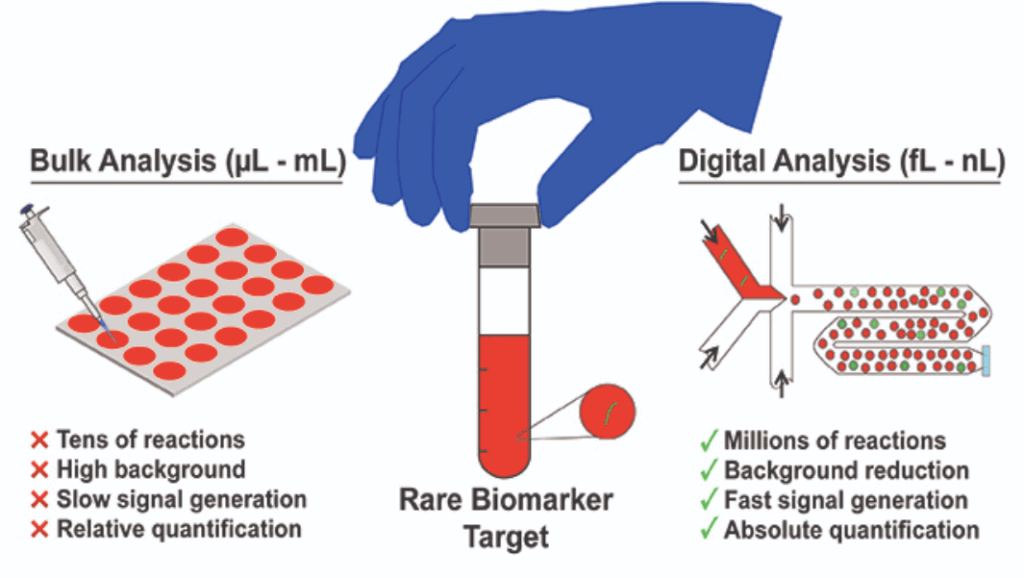
Droplet encapsulation is particularly well-suited to improve the outcomes of high-throughput screening because it allows for the generation of highly monodisperse droplets, which provides homogeneous reaction conditions. These reactions happen in the nL- and pL-range, reducing reagent consumption, and the droplets can be created with frequencies up to 10 kHz, allowing high-throughput screens of up to 100 million reaction conditions per day [12].
Some applications of droplet-based high-throughput screening are outlined in the sections below.
Single-cell analysis is optimized in droplet-based microfluidics
The advent of droplet-based microfluidics and droplet encapsulation allows biologists to screen for heterogeneity in cell populations through single-cell proteomics and genomics.
In single-cell proteomics, cells are confined in a minimal volume, which makes most biomarkers almost readily detectable. Either by cell secretion or cell lyses within the droplet, the detection of molecules of interest is usually done by fluorescence-labelled antibodies, providing a “0 or 1”-like result, which gives the name of digital microfluidics for this kind of analysis. There is a significant interest in the field to make the results quantifiable, opening even more possibilities for using droplets [1].
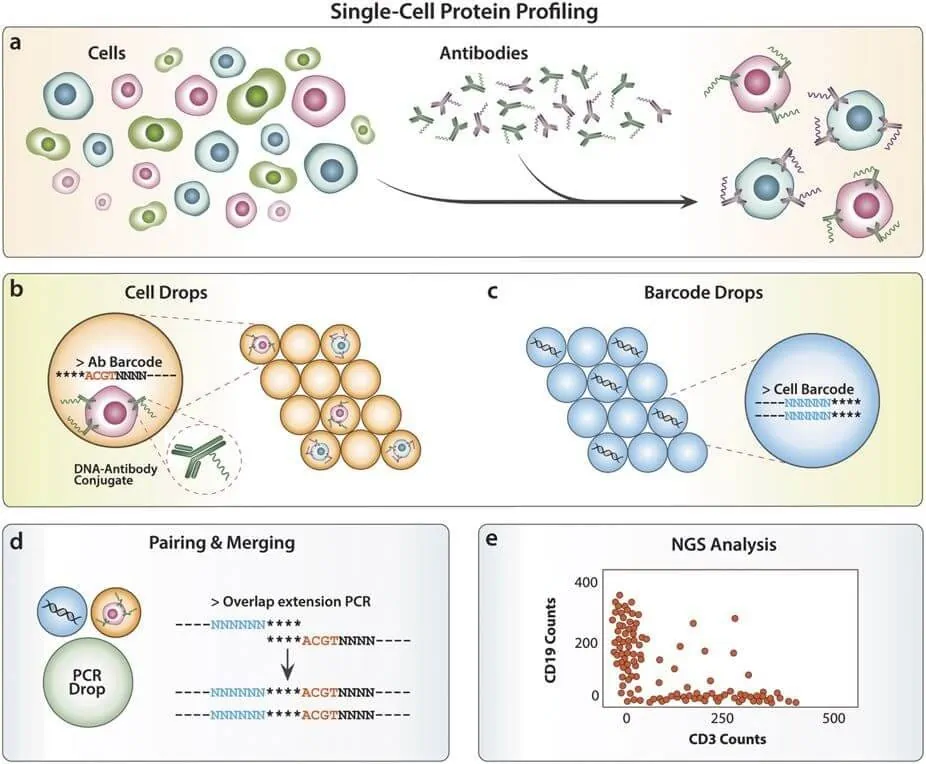
Single-cell genomics allows for DNA targeting by single-cell encapsulation with primer-functionalized beads. Therefore, it is possible to map the genomes of entire cell subpopulations with droplets. In this line, there is a commonly used sequencing protocol for RNA called Drop-seq, one of the few applications with commercial exploitation in this field to date [14].
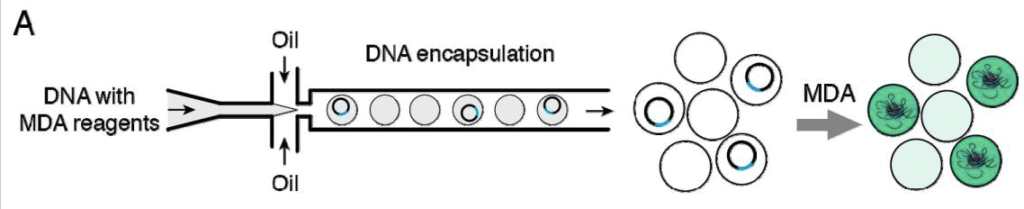
Droplet encapsulation enables ultrahigh-throughput in directed evolution
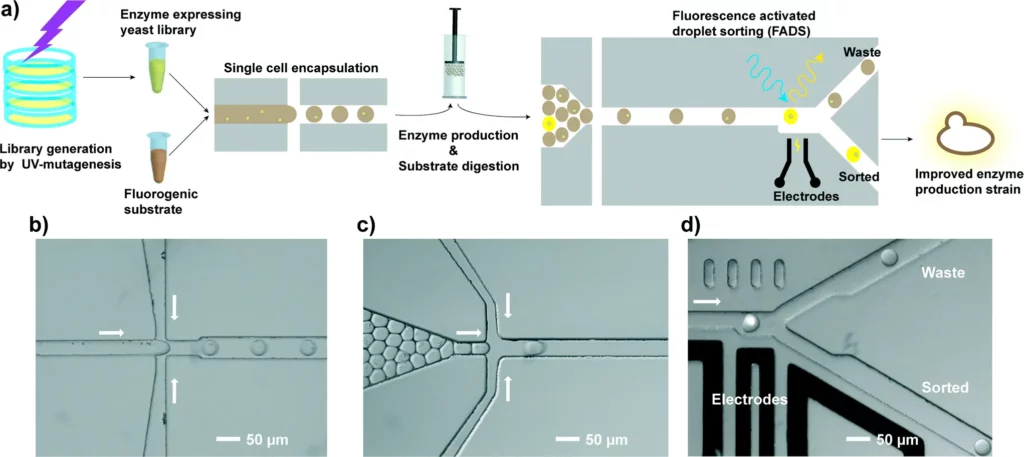
Directed evolution is a process that confines and speeds up the evolution of proteins by mimicking natural selection in vitro. It has been mainly applied to select and fine-tune interesting enzymes for industrial use.
The miniaturization, improved control and homogeneity, and sorting capabilities of droplet-based microfluidics allow researchers to develop ultrahigh-throughput directed evolution (over 106 compounds per day). These systems enabled the screening of 108 proteins, which exceeds the capabilities of standard approaches. For example, in a conventional plate assay, this experiment would take two weeks to screen only 2000 proteins [11].
Droplet-based drug delivery systems
Besides being microreactors, droplets can be transformed into drug delivery systems (DDS). The droplet encapsulation methodology is similar to the ones described above, with the drug being diluted in the dispersed phase and its concentration being controlled by the flow rate.
However, depending on the administration route of the medication, the droplets might need to be turned into microparticles/nanoparticles through a solidification step. Also, some droplet-based DDS might have their solutions functionalized to respond to a specific pH or temperature, acting as controlled delivery systems [16].
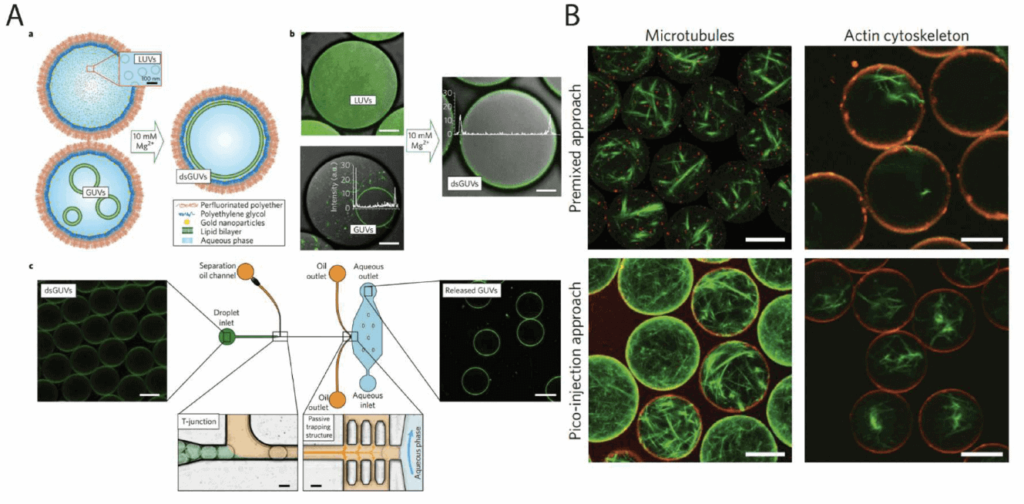
Droplets can be templates for artificial cells
Droplet compartmentalization can be used as a template for artificial cells. Depending on the research goal, it can be used as a membrane-free compartment for isolated biochemical reactions mimicking specific cell functions or as a template for the creation of lipid bilayers.
More complex designs allow the encapsulation of droplets inside larger droplets to mimic the compartmentalization of natural cells and to provide better timely control of the reactions since the reagents are not all mixed in the dispersed phase before droplet formation. Majumder et al., 2019 [17] recently reviewed how droplets can generate artificial cells.
Key takeaways from droplet encapsulation
Droplet encapsulation provides the ability to create microreactors in a highly controlled manner and in large quantities in the form of monodispersed droplets, which affords clear advantages to applications in biological research. Also, microfluidics setups are flexible and can be adjusted to fit several applications, which explains their rapid adoption outside the physics and fluidics fields.
For researchers who do not have a microfluidics background, the task of setting up the first microfluidics experiment might seem daunting. However, several solutions are aimed at turning microfluidics into an easy-to-use tool that can be employed without previous expertise. For example, an easy droplet-generator pack, and an automatic droplet size regulator pack are available for researchers interested in including droplets in their current research without learning about an entirely new field.
Review done thanks to the support of the Protomet H2020-MSCA-ITN-2018-Action “Innovative Training Networks”, Grant agreement number: 813873
Author: Camila Betterelli Giuliano, PhD
Partnership[at]microfluidic.fr



References
- N. Wen, Z. Zhao, B. Fan, D. Chen, D. Men, J. Wang and J. Chen. Development of Droplet Microfluidics Enabling High-Throughput Single-Cell Analysis. Molecules, vol. 21, 7, 2016.
- T. Schneider, J. Kreutz and D. T. Chiu. The Potential Impact of Droplet Microfluidics in Biology. Analytical Chemistry, vol. 85, 7, 2013.
- Zubaite, G., Simutis, K., Galinis, R., Milkus, V., Kiseliovas, V., & Mazutis, L. Droplet Microfluidics Approach for Single-DNA Molecule Amplification and Condensation into DNA-Magnesium-Pyrophosphate Particles. Micromachines, 8(2), 62, 2017.
- L. Shang, Y. Cheng, and Y. Zhao. Emerging Droplet Microfluidic. Chemical Reviews, vol. 117, 2017.
- T. S. Kaminski, O. Scheler and P. Garstecki. Droplet microfluidics for microbiology: techniques, applications and challenges. Lab on a Chip, vol. 16, 2016.
- B. B. Wang, X. D. Wang, T. H. Wang and W.M. Yan. Electrocoalescence behavior of two identical droplets with various droplet radii. Applied Thermal Engineering, vol 111, 25, 2017.
- M.Sesen, T. Alan and A. Neild. Microfluidic on-demand droplet merging using surface acoustic waves. Lab on a Chip, vol. 14, 2014.
- J. Sivasamy, Y. C. Chim, T. N. Wong, N.T. Nguyen and L. Yobas. Reliable addition of reagents into microfluidic droplets. Microfluidics and Nanofluidics, vol. 8, 3, 2010.
- H. Yuan, Y. Pan, J. Tian, Y. Chao, J. Li, H. C. Shum. Electricity-free picoinjection assisted droplet microfluidics. Sensors and Actuators B: Chemical, vol 298, 2019.
- A. R. Abate, T. Hung, P. Mary, J. J. Agresti and D. A. Weitz. High-throughput injection with microfluidics using picoinjectors. PNAS, 107 (45), 2010.
- F. W. Y. Chiu and S. Stavrakis. High‐throughput droplet‐based microfluidics for directed evolution of enzymes. Electrophoresis, 2019
- L. D. van Vliet and F. Hollfelder. Microfluidic Droplets and Their Applications: Diagnosis, Drug Screening and the Discovery of Therapeutic Enzymes. IFMBE Proceedings, 2019.
- P. Shahi, S. C. Kim, J. R. Haliburton, Z. J. Gartner & Adam R. Abate. Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Nature Scientific Reports, 7, 2017.
- E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev and S. A. McCarroll. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets, Cell, vol. 161, 5, 2015.
- S. L. Sjostrom, Y. Bai, M. Huang, Z. Liu, J. Nielsen, H. N. Joenssona and H. A. Svahn. High-throughput screening for industrial enzyme production hosts by droplet microfluidics. Lab on a chip, issue 4, 2014.
- H. Feng, T. Zheng, M. Li, J. Wu, H. Ji, J. Zhang, W. Zhao and J. Guo. Droplet-based microfluidics systems in biomedical applications. Electrophoresis, vol. 40, 11, 2019.
- S. Majumder, N. Wubshet and A. P. Liu. Encapsulation of complex solutions using droplet microfluidics towards the synthesis of artificial cells. Journal of Micromechanics and Microengineering, vol. 29, 8, 2019.
- Weiss, M., Frohnmayer, J., Benk, L. et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nature Mater 17, 89–96 (2018)
Check the other Reviews
FAQ - Droplets encapsulation for biological applications: a review
What is the actual meaning of the concept of droplet encapsulation in biology?
Practically, it refers to the practice of immiscible biological sample (cells, enzymes, beads, DNA, reagents etc.) into large volumes of small, isolated water-in-oil compartments (usually droplets, often a few micrometers across, created in a microfluidic chip). Every drop is a repeatable mini-reactor: identical volume, identical chemistry, time-to-time. The large-scale payoff is that you no longer average everything in bulk, that you are now able to consider the issue of heterogeneity cell-to-cell variability, rare events, the issue of outliers that are of more importance than the mean.
So what is the difference with miniaturization to the microscale? And a droplet is no more than a tiny tube?
In the microscale the surface/volume ratio is significantly larger which typically results in rapid mixing and rapid heat transfer. And due to the fact that the droplets are monodisperse (nearly identical), you are able to carry out reactions under truly similar conditions. That is why people refer to droplets as standardized microreactors and not miniature test tubes.
How would you most simply wrap a single cell in a droplet- and why is it surprisingly inefficient?
Simple dilution is the default approach: you dilute the cell suspension during the dispersed phase, and then make the droplets and hope the statistics such that the vast majority of the droplets will contain no cells or one cell. The sting is the Poisson loading penalty: the likelihood that each droplet will contain a single cell will be approximately 36.8, in other words there is a significant proportion of empty droplets in a collection. When you require two distinct entities in one (e.g. one cell and one bead, or two cell types), the probability may fall as low as about 13.5% for an ideally droplet, hence you consume sample, time and analysis bandwidth.
In case the process of dilution is inefficient, what do even smarter people do in terms of encapsulation?
Multiple aqueous inlets (co-flow style): two (or more) dispersed streams are pinched off to create a droplet and are mixed, useful when it is important when the mixing takes place.
Droplet- droplet fusion: droplets are made individually (some with a different reagent) and fused later. Electro-coalescence (upsetting stabilization of the droplets by an electric field) or other methods like acoustic tweezing can be used to cause fusion and coalesce the droplets together.
Addition of reagents after formation: you do not make all the decisions in advance, but inject into droplets that have been previously formed. That naturally results in plug-based injection or picoinjection.
What is picoinjection and when is it worth the bother?
Picoinjection is simply fine-tuning dosing in miniature amounts, normally via an actuated electric field injecting minute amounts, as small as sub-picoliters of control, of droplets as they migrate in oil. It’s worth it when you need to:
-deliver a reagent at a particular time (not at drop birth),
-measure titrate concentrations in droplets without resettling the emulsion,
-mix multi-step tests in which order of mixing is significant (enzyme kinetics, sequential labeling, timed lysis etc.).
It requires more of the chip design and instrumentation, however, it provides the experimental freedom that you just cannot get with one-shot encapsulation.
In what are the brightest areas of droplet encapsulation in biology?
High-throughput screening (HTS): typical HTS is provided at about 104-105 tests/day in well plates and robots; droplet platforms may be massively scaled, since droplets can be made at frequencies of 10 kHz. Screening claims in optimized nL-pL reaction volumes and continuous flow can achieve reaction conditions of about 100 million/day.
Single-cell assays: here you cease to average signals, and begin reading cell-to-cell variation, and may have a feel of that which is digital (signal/no signal) and can be measured well with good calibration.
Directed evolution: Since rare high-performers are selected by sheer numbers, droplet sorting and miniaturization can go to >106 variants/day, and screens of the order of 108 proteins have been discussed, far more than is practicable on plates (where even 2,000 variants might require >two weeks with a standard assay).
And what are the droplets themselves as a template to artificial cells (here an artificial cell is anything, is it not)?
It is a large umbrella called an artificial cell. In this respect the droplets are employed as:
-membrane-free isolated biochemical reaction compartments that simulate particular cell-like activities, or
-lipid bilayer formation templates, in which droplets assist in production of vesicle-like objects (such as giant unilamellar vesicles).
More complicated architectures–such as droplets within larger droplets–can recapitulate the compartmentalization of actual cells and provide increased temporal control (since not all things need to be mixed prior to the formation of droplets).
What are the key bottlenecks in the attempt of a biology lab to use droplets the first time?
Scientific aspect is not always the most difficult–the apparatus. Some of the areas of common friction are:
-stabilization of emulsions (surfactants, choice of oil),
-regulating the droplet size and maintaining it in the long-run,
-balancing Poisson loading and cost of sample,
-combining incubation, merging/injection and detection and not transforming the experiment into a plumbing project.
That is the reason why commercially available products (a simple droplet generator, or an autofocusing droplet size controller) can be a convenient point of entry of a group that seeks output and does not aspire to become a microfluidics professional.
In which stages does a microfluidics SME contribute in a Horizon Europe-type project (not just we can make chips)?
Droplet microfluidics is the layer that has the power of making or breaking the biology scalable and reproducible, the enabling layer of innumerable consortia. A committed SME has the capability to contribute something that academic partners may not be able to maintain over long term:
-setting up the entire experimental system (fluidics, control, interfacing, repeatability),
-rapid plasticity and experimentation of chips and fixtures,
-converting a lab demonstration into something that is substantial enough to be multi-site teste.
We are habitually the microfluidic engineering partner in European consortia and we, too, assist in writing proposals such that the microfluidics component is plausible, verifiable, and the de-risking of the microfluidics element is plausible. When tracking proposal outcomes in our internal experience tracking proposal outcomes, an early embedded microfluidics SME may positively increase the competitiveness of the proposal, with differences as high as a doubling effect over control success rates, since reviewers can observe execution capacity, but not merely intent.