Microfluidics drug discovery and biomedical research: Emerging technologies and applications
Author
Emma Glickman
Publication Date
Keywords
Drug Discovery
disease modeling
Organ-on-a-chip
Pharmaceutical Advancement
Droplet Microfluidics

Need advice for your biomedical research?
Your microfluidic SME partner for Horizon Europe
We take care of microfluidic engineering, work on valorization and optimize the proposal with you
Introduction to microfluidics drug discovery and biomedical research
Drug discovery and biomedical research are central to advancing global health by improving therapeutic development, reducing disease burden, and preparing for emerging public health challenges. Traditional drug screening and biomedical research methods, including static cell-culture and animal models, remain costly, low-throughput, and poorly scalable. These systems often lack the physiological complexity or relevance necessary to accurately predict human responses, contributing to high rates of late-stage drug attrition. Nearly 90% of drugs that succeed in animal studies later fail in human trials due to adverse or ineffective responses [1].

Likewise, disease mechanisms that appear well characterized in animal models frequently translate poorly to human physiology [2]. While conventional in vitro and in vivo approaches remain valuable for early-stage hit identification and basic disease modeling, their limited physiological fidelity constrains their capacity to reproduce key aspects of human biology. As a result, traditional cell-culture and animal-based workflows are generally slow and low-throughput, leading to lengthy preclinical development cycles, a critical limitation when disease progression and global health threats can evolve far more rapidly than current research timelines allow [3].
These inefficiencies underscore the urgent need for high-throughput, cost-effective, and physiologically relevant drug screening and biomedical research platforms. Emerging technologies, such as droplet microfluidics, microfluidic-based organ-on-chip (OoC) and lab-on-chip (LoC) systems, offer unique potential. These devices provide precise control over cellular microenvironments, minimize reagent consumption, and enable parallelized, automated experimentation. Recent advances in materials science, fabrication techniques, and integration with standard laboratory devices have driven a shift from proof-of-concept devices towards robust, reproducible platforms.
By bridging the gap between experimental control and biological relevance, microfluidic systems present a powerful alternative to conventional screening approaches. They hold the promise of modeling complex human diseases and tissue responses, improving predictive power, and accelerating drug discovery and biomedical advances. This review examines recent advances in microfluidics drug discovery and biomedical research using droplet microfluidics, OoCs, and LoCs. Emphasis is placed on their contributions to high-throughput capacity, low reagent usage, and physiological fidelity, highlighting how these technologies are transforming preclinical testing and disease modeling into a more predictive, scalable, and human-relevant process.
Emerging technologies in microfluidics drug discovery and biomedical research
Microfluidic platforms have become integral tools in modern drug development and biological research. Key among these are droplet microfluidics, organ-on-chip (OoC) systems, and lab-on-chip (LoC) devices. These technologies are emerging as key approaches to bridge experimental precision with physiological relevance.
Droplet microfluidics
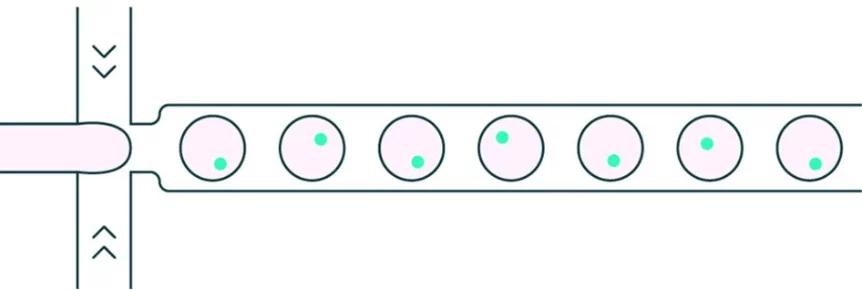
Droplet microfluidics is a technology that generates and manipulates highly uniform droplets on the femtoliter-to-nanoliter scale. By flowing two immiscible liquids through microfluidic channels, droplets are produced as independent microreactors that can encapsulate cells, biomolecules, or drugs. This approach enables massively parallel high-throughput screening at dramatically reduced reagent costs.
In microfluidics drug discovery, each droplet serves as a miniaturized assay chamber, enabling precise control over drug concentrations, exposure conditions, and combinatorial treatments. The small volume accelerates reaction kinetics and data acquisition and reduces reagent costs, while droplet uniformity ensures reproducibility [5].
Organ-on-a-chip
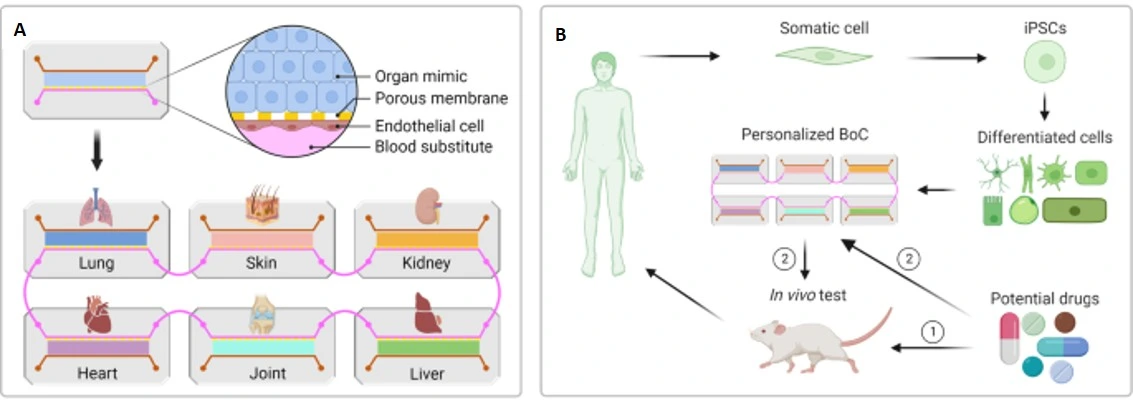
Organ-on-a-chip (OoC) platforms are microengineered devices that recapitulate a human organ at the microscale. OoCs integrate fluid flow, mechanical forces, and multicellular co-culture to better recapitulate physiological responses than static in vitro models. Importantly, OoCs provide a more predictive platform for biomedical research and microfluidics drug discovery by modeling diseases, drug absorption, metabolism, and toxicity.
OoCs enable researchers to study human-relevant cell types, often in a 3D structure designed to mimic human tissue physiology. The incorporation of multiple cell types, patient-derived cells, or disease-specific cells enables precision and even personalized drug development. OoC co-cultures have been developed that accurately model many human organs, including the heart, kidney, lung, gut, liver, and even brain. Single-organ OoCs allow precise modeling of organ-specific toxicity or adverse effects [7].
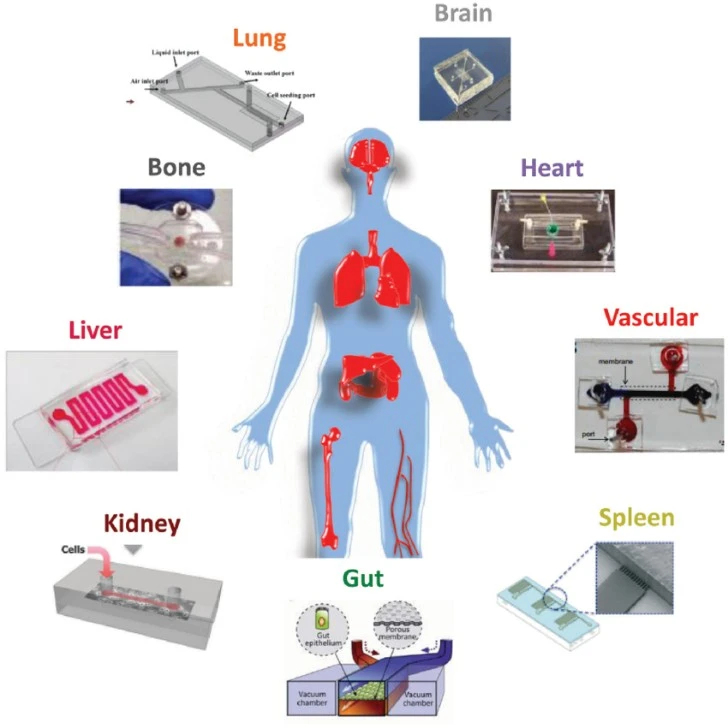
The connection of multiple OoCs featuring different tissues gives researchers the ability to assess systemic drug responses and model how a drug will undergo absorption, distribution, metabolism, and excretion (ADME) in a human-relevant platform [7].
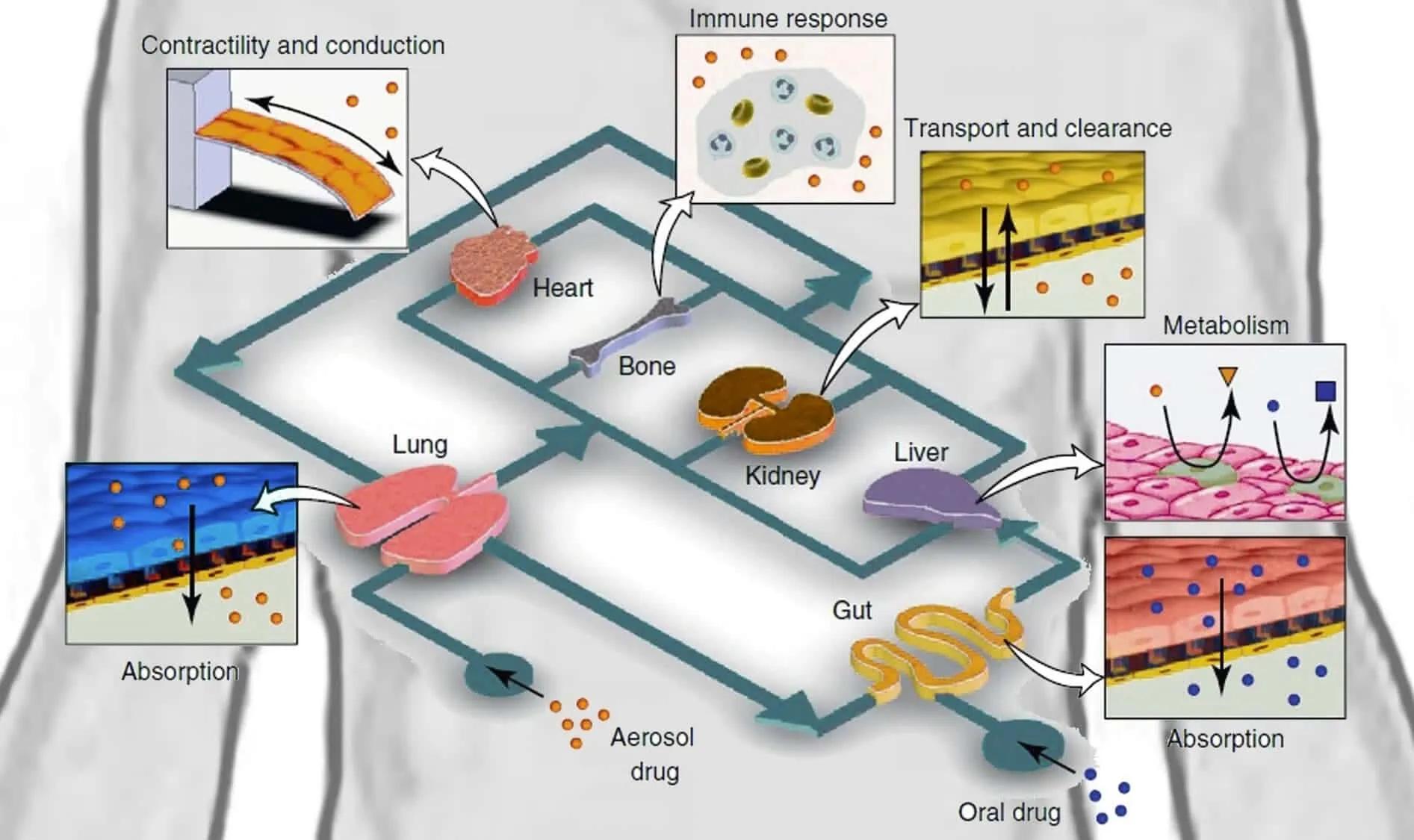
OoCs, when used in microfluidics drug discovery and biomedical research, enable researchers to thoroughly test drug candidates in systems that are more relevant to human physiology, healthy or diseased, than standard culture systems and animal models. Use of OoCs can drastically reduce late-stage failures by more accurately predicting a drug’s human response. This capability makes them an important alternative to traditional preclinical drug screening methods, such as static 2D cell culture.
Lab-on-a-chip
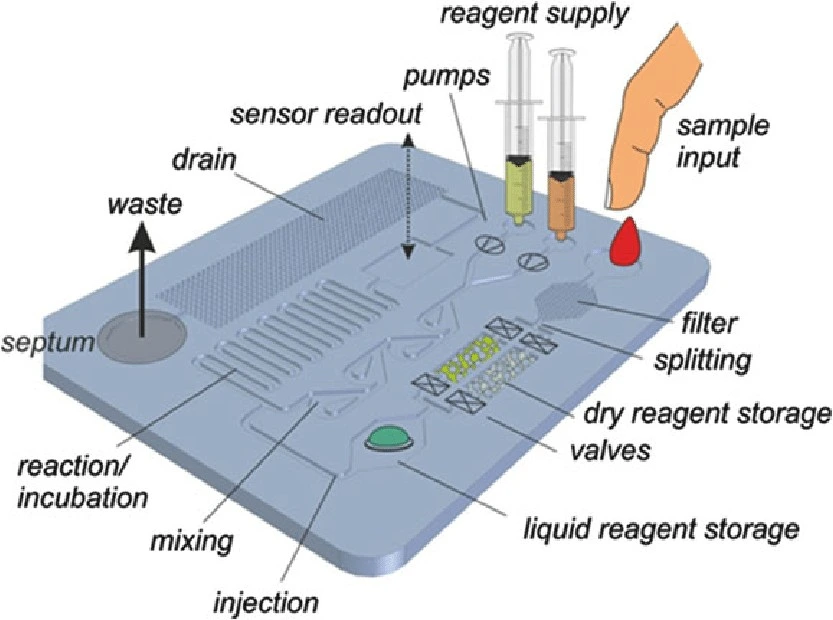
Lab-on-a-Chip (LoC) platforms are microengineered devices that integrate and miniaturize multiple laboratory processes onto a single chip. They automate complex biological and analytical processes, enabling precise fluid control and rapid analysis.
LoCs utilize microfluidics to integrate sample preparation, reactions, and detection into a single chip. They can handle extremely small reagent volumes, resulting in lower reagent costs and less waste. The miniaturization of LoCs creates short diffusion distances, rapid heating, and high surface-area-to-volume ratios, enabling rapid reactions and analysis while enabling precise environmental control. Their compact size also enables massive parallelization, creating the option for ultra-high-throughput analysis [10].
LoC devices offer a range of use cases, from point-of-care diagnostics to high-throughput drug screening. Their capacity for automation, miniaturization, and parallelization allows for quick and efficient drug candidate screening, ultimately reducing the time and cost of preclinical research and improving throughput of microfluidics drug discovery and biomedical research.
Droplet microfluidics, OoCs, and LoCs represent major advancements in microfluidics drug discovery and biomedical research. These technologies can be applied to a wide range of applications, from high-throughput drug screening to precision medicine and disease modeling. The development and implementation of these technologies will greatly alter the drug development and biomedical research workflows.
Emerging applications in microfluidics drug discovery and biomedical research
Microfluidic technologies have found diverse and rapidly expanding applications across pharmaceutical and biomedical research. Here, some examples are described across key domains, including functional readouts, high-throughput compound screening, pharmacokinetics and pharmacodynamics, disease modeling, and precision medicine.
Functional readouts
Basic aspects of cellular health and functionality are crucial for understanding drug responses and selecting the best candidates during drug screening. Understanding how a compound affects a cell’s morphology, metabolism, barrier integrity, transport mechanisms, and DNA/RNA/protein expression can provide direct insight into a cell’s or tissue’s health and the safety and efficacy of a drug of interest. Integration of these functional readouts into a high-throughput droplet, OoC, or LoC platform provides researchers with a quick, comprehensive, and physiologically relevant understanding of a compound’s impact on a cell or tissue, advancing both microfluidics drug discovery and biomedical research.
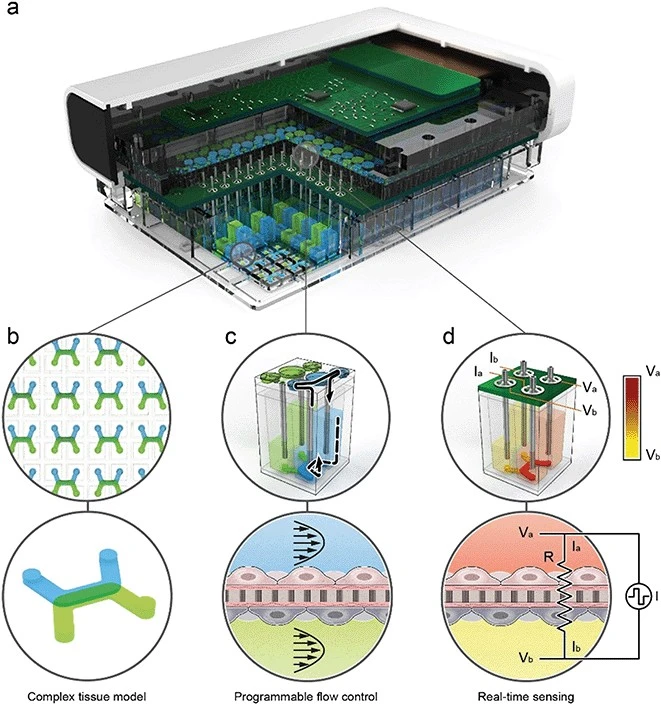
Recently, Azizgolshani et al designed a high-throughput LoC platform integrating programmable fluid flow, real-time sensing, and compatibility with automated imaging systems. Their device supports trans-epithelial electrical resistance (TEER) analysis, oxygen consumption monitoring, and active transport assays, while generating over 10,000 high-resolution images in under one hour. The system also enabled the extraction of high-quality RNA for transcriptomic profiling of drug responses. By combining parallelized assays, real-time monitoring, and multi-organ compatibility, their platform demonstrates the potential for scalable, cost-effective, and physiologically predictive drug testing [12].
High-throughput compound screening
Current drug discovery pipelines rely on static in vitro and animal models. These methods are expensive, require lengthy development timelines, and often fail to accurately predict human responses. These issues highlight the need for efficient, large-scale testing of chemical compounds.
High-throughput screening (HTS) refers to automated, miniaturized processes that enable rapid testing of thousands to millions of compounds against specific targets. HTS enables parallelization and the use of small sample sizes, accelerating the identification of drug candidates by drastically reducing experimental time and reagent use. In the context of microfluidics-based drug discovery, droplet microfluidics and high-throughput OoC devices address the bottlenecks posed by traditional screening methods and bridge critical gaps in translational biomedical research.
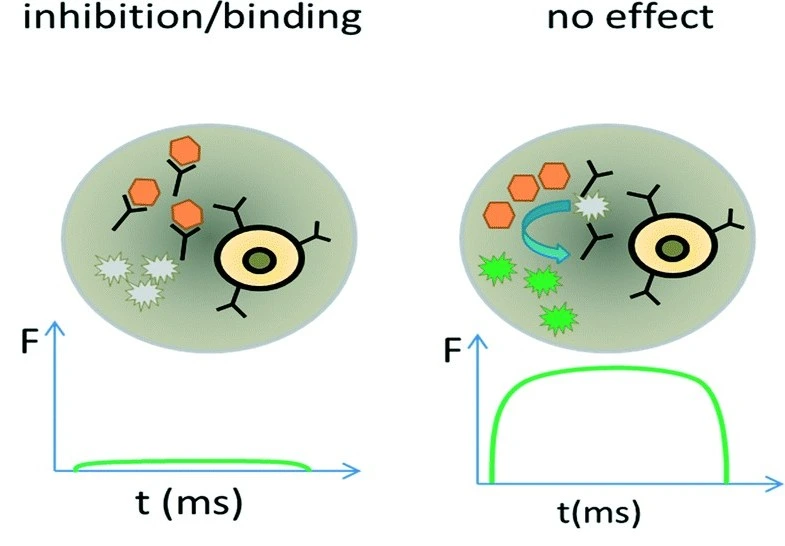
Shembekar et al. implemented droplet microfluidics for high-throughput screening of antibodies that inhibit angiotensin-converting enzyme 1 (ACE1), a major drug target for the treatment of hypertension and congestive heart failure. They encapsulated single hybridoma cells expressing ACE1 targets into 660 pL droplets, yielding antibody concentrations of ~20 ug/mL within just 6 hours, much faster than in traditional hybridoma cultures. After incubation, droplets were mixed with fluorogenic substrates to detect inhibitory antibodies. Remarkably, more than 300,000 clones were screened in a single experiment, yielding high accuracy with 17 out of 18 selected cells producing functional antibodies [13].
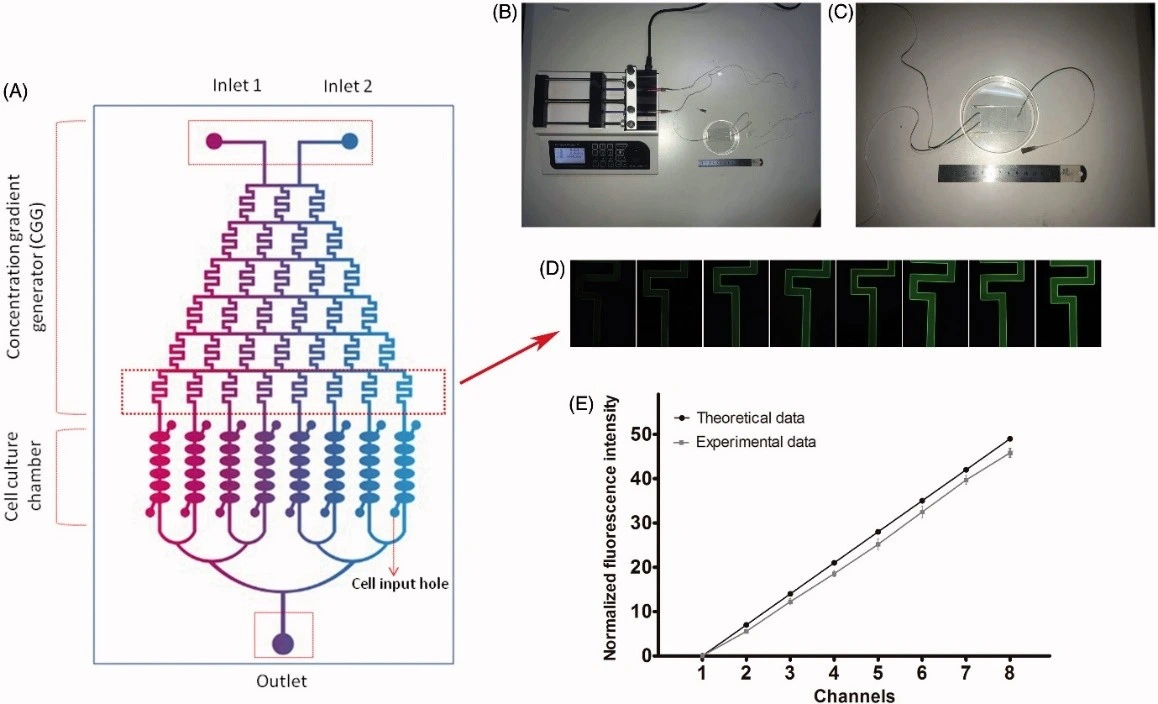
Ming et al developed a cartilage-on-chip device to investigate the effects of resveratrol on chondrocytes. Their system incorporated a concentration gradient generator connected to eight parallel culture chambers, each subdivided into six compartments. This design enabled simultaneous testing of multiple drug concentrations with high fidelity, rapidly identifying optimal scaffold-doping doses to promote cartilage repair. Compared to conventional dilution-based assays, this platform provided results that more closely reflected human physiology and required far less labor [14].
Pharmacokinetics and pharmacodynamics modeling
Pharmacokinetics (PK) and pharmacodynamics (PD) are essential in understanding how drugs behave in the body. PK refers to the absorption, metabolism, and secretion of compounds, and PD characterizes biochemical and physiological effects. In the fields of microfluidics drug discovery and biomedical research, accurate modeling of human physiology enables a better understanding of PK and PD responses to drugs. Use of human-relevant models, such as OoC systems, provides improved predictions of PK and PD compared to traditional static cultures or animal models. Recapitulating tissue-tissue interactions, dynamic flow conditions, and metabolic linkages allows researchers to study real-time PK/PD relationships and predict drug responses in a controlled environment.
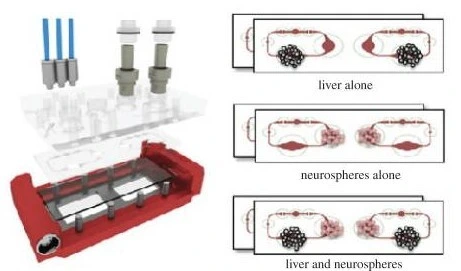
Herland et al. developed a ‘first-pass multi-organ-chip-system’ to quantitatively predict human pharmacokinetic responses to drugs. They used automated liquid transfer to link gut/bone marrow, liver, and kidney chips to a reservoir containing a blood-substitute medium. Each organ chip included an epithelial and endothelial layer. The group administered oral nicotine in the gut/liver/kidney model and intravenous cisplatin in the bone marrow/liver/kidney model.
The experimental results from the chip systems were computationally scaled to match human clinical data and successfully demonstrated in vitro-to-in vivo translation in the absorption, distribution, and clearance parameters to an extent not previously modeled in vitro. Importantly, the chip systems also accurately displayed toxicities observed in vivo, such as neutropenia and anemia induced by cisplatin, strengthening the relevance of OoC platforms in translational biomedical research [15].
Maoz et al. modeled the pharmacodynamic responses of human neurovascular cells to methamphetamine using a microfluidically linked human neurovascular unit-on-a-chip (NVU chip). The group connected two blood-brain barrier (BBB) chips on either side of a brain chip to mimic the influx and efflux pathways of molecules across the BBB, into, and out of neural tissue. The arrangement was designed to mimic the physiological compartmentalization of the human NVU. The chip system showed that fluidic linkage significantly altered protein expression and paracrine signaling compared to unlinked cells. To further assess the NVU system, the researchers challenged their chips with methamphetamine and discovered that acute meth treatment transiently and reversibly increases BBB permeability, temporarily allowing large molecules to pass through the BBB, only in the presence of the drug and only on the BBB chip representing influx to the brain.
These results closely matched animal and clinical observations of the impact of meth on the BBB. It was also found that endothelial and perivascular cells supplied metabolic substrates, such as pyruvate, lactate, and glutamine, to the brain chip, thereby directly influencing neurotransmitter synthesis. The metabolic coupling, altered protein expression, and paracrine signaling, and close match to clinical data indicate the importance of linked OoC systems in drug discovery and highlight their potential as replacements for animal or clinical testing in biomedical research [17].
Disease modelling
Disease modeling is an important aspect of drug discovery. Testing drug candidates on healthy cells or healthy patients limits understanding of drug response to cases where the patient is already healthy. On the other hand, unhealthy patients are less likely to be able to donate cell samples or undergo clinical trials. Additionally, while animal disease models often offer valuable insights into human drug responses, the inherent physiological differences can lead to drug failures in clinical trials, even after drugs have passed animal trials. Modeling diseases in vitro using human-relevant platforms is a powerful application of microfluidics drug discovery and biomedical research, as it improves drug response prediction and supports the development of more translatable therapies.
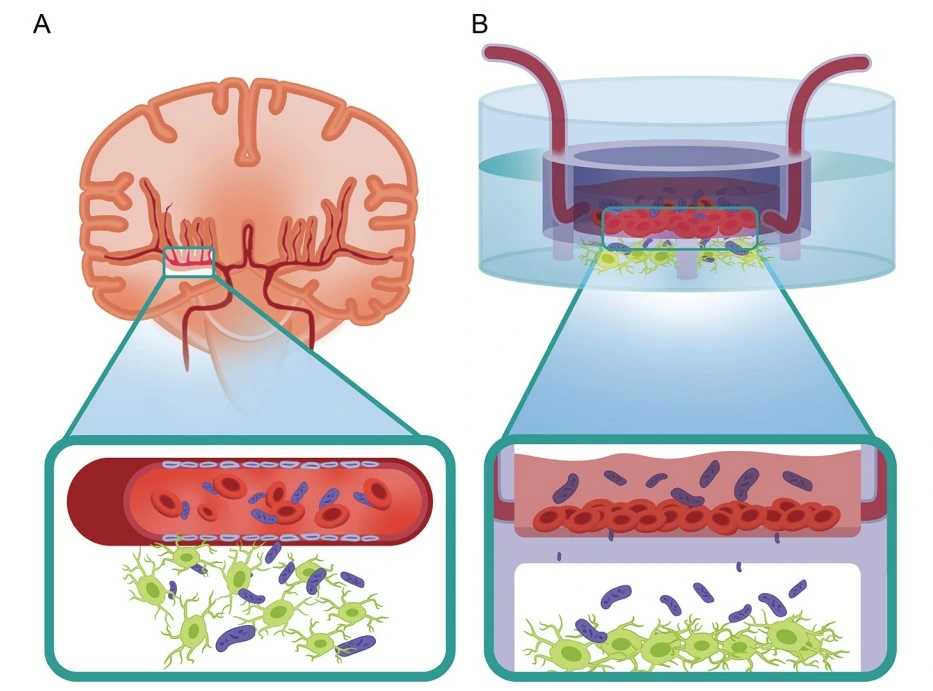
Rauti et al. developed a neurovascular unit (NVU) chip with microfluidically induced flow to study the pathophysiology of newborn bacterial meningitis (NBM), specifically the pathway of Escherichia coli across the BBB and its effects on the brain. The NVU chip features human umbilical vein endothelial cells (HUVEC) cultured in the apical compartment and primary rat neurons in the basal compartment. The apical compartment was cultured under flow, maintained by microfluidic connections to pumps. Experiments were conducted in monoculture and co-culture conditions, first assessing the individual endothelial and neuronal responses to infection, then the effect in combined co-culture in the NVU chip, with bacteria initially introduced only to the apical compartment.
The endothelial mono-culture showed significant BBB disruption resulting in increased permeability and pronounced inflammatory responses. The neuronal monoculture showed reduced protein volume and decreased spontaneous electrical activity, indicating a loss of network function. The combined co-culture displayed the same characteristic responses, but neuronal impact was delayed by the BBB, more closely mimicking the progression of infection seen in vivo.
The bacterial strains tested on the NVU chip carried group 4 O-antigen capsules rather than the classic K1 capsule typically found in E. coli strains causing NBM. The inflammatory and functional injury of cells in response to infection with E. coli strains lacking K1 capsules has important implications for therapeutic development that has typically targeted only K1-capsule E. coli strains. This discovery highlights how physiologically relevant disease models can advance biomedical research and uncover new therapeutic and vaccine targets [19].
Precision medicine
Precision medicine is an innovative healthcare approach that tailors treatment to the individual rather than following a “one-size-fits-all” model. It draws on a patient’s genetics, lifestyle, and environment to guide medical decisions and therapy selection. In the lab, this often involves testing drugs or treatment protocols directly on a patient’s cells to identify the most effective option quickly, minimizing patient exposure to ineffective therapies. However, this strategy is limited by the availability of patient-derived samples and the rapid progression of some diseases, which demand rapid analysis with minimal sample sizes. Microfluidic platforms address these challenges by enabling miniaturization and parallel testing, making them crucial tools for advancing both precision medicine and microfluidics drug discovery in modern biomedical research.
Zhai et al developed a digital microfluidic (DMF) LoC platform for drug screening using primary patient tumor cells. The system allows parallel screening of multiple drugs on cells obtained from biopsy samples. It operates through an array of electrodes embedded in a glass surface that manipulate droplets containing cells and drugs via electrical signals. By applying electrode-sharing logic, the design minimizes the number of electrodes needed, enabling a compact, efficient layout. The chip measures only 4×4 cm^2 and is housed within a 23x16x3.5 cm^3 control unit. Its small scale allows experiments to run with hundreds to thousands of cells and very low reagent volumes, significantly reducing costs.
The platform supports parallel testing of up to three drugs. After incubation, fluorescent microscopy is used to assess cell viability and generate dose-response curves. The entire workflow is completed within 36 hours, limiting cellular expansion and maintaining the integrity of dose-response profiles. The device was validated using both animal models and patient samples, showing a strong correlation between in-chip drug effectiveness and clinical outcomes. Furthermore, the chip can be reused up to ten times, providing an additional cost advantage [20].
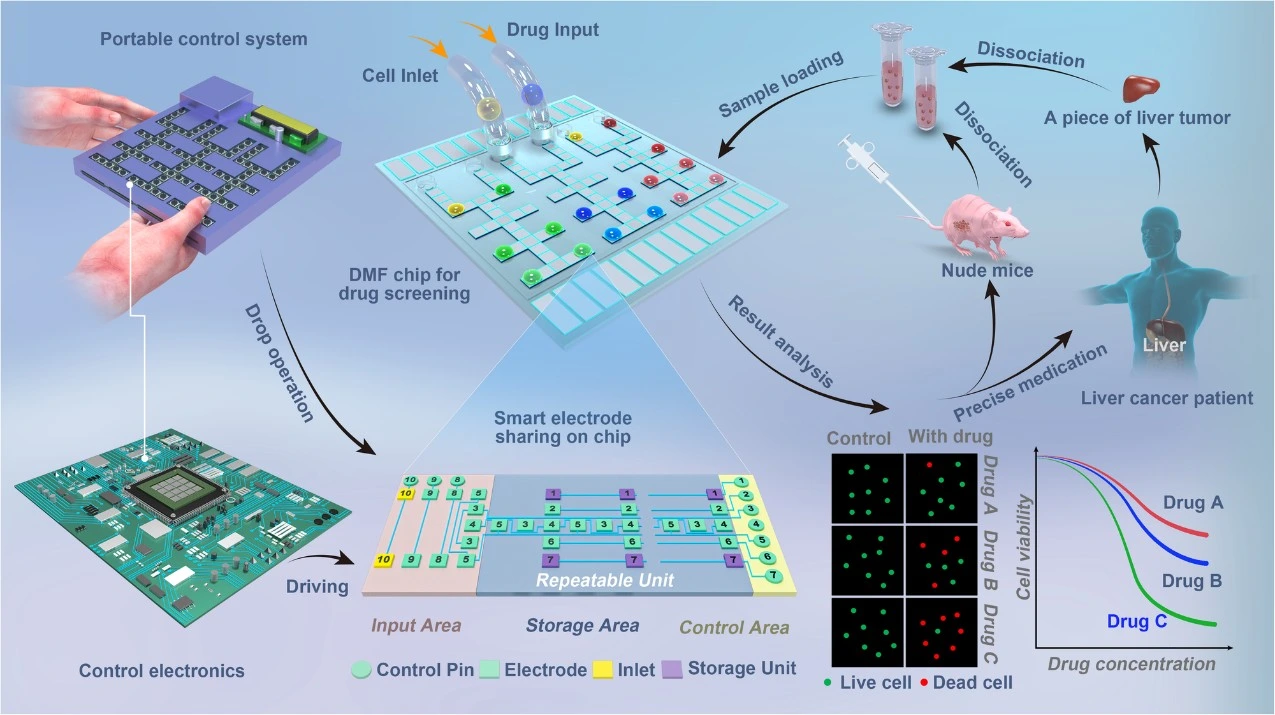
Moll et al. developed a microtiter microfluidic plate for high-capacity cell culture and precision treatment that complies with ANSI/SLAS standards for laboratory automation. The system uses compartmentalized cell cultures (CCCs), in which cell populations are physically separated within a single device via microfluidic channels and membrane barriers. This setup enables precise control over local microenvironments. However, conventional CCC systems often have limited capacity, rendering them unsuitable for large-scale pharmaceutical applications.
To overcome these limitations, Moll et al. created the microchannel microtiter plate (MC-plate). The device consists of three layers: a standard-layout 384-well plate on top, a central microfluidic layer that connects the chambers through microchannels, and a clear sealing film on the bottom. Each CCC unit features three open upper compartments linked by microchannels, yielding a total of 96 experimental units per plate. Because its upper layer follows a standard format, the MC-plate is compatible with automated liquid handling systems [21].
The novelty of this system lies in its microchannel network, specifically designed for neurological studies. The microchannel dimensions allow only long neuronal axons to extend between compartments, effectively physically separating cell culture compartments from one another. Chemical isolation is accomplished through hydrostatic pressure differences. Treated compartments receive 50 μL of liquid, and the adjacent compartments receive 100 μL, resulting in ~5 mm of height difference that drives directional flow and prevents compound crossover. The researchers demonstrated stable fluidic isolation for at least 72 hours, successfully confining fluorescent molecules and viral vectors to designated compartments.
The MC-plate supported the culture of both mouse primary neurons and human induced pluripotent stem cell (IPSC)-derived neurons for up to three weeks. Functional connectivity was confirmed through spontaneous electrical activity and synchronized calcium signaling, both key indicators of neuronal communication. With its scalable design, fluidic precision, and compatibility with automation, the MC-plate provides a powerful platform for studying neuronal disease mechanisms, protein aggregation, and cell-cell signaling, making it important in the development of drugs for neurological diseases and a valuable resource for biomedical research.
Advantages and limitations in microfluidics drug discovery and biomedical research
Microfluidic technologies like droplet microfluidics, OoCs, and LoCs have transformed experimental workflows in drug discovery and biomedical research through unprecedented miniaturization, integration, and automation. This section introduces the advantages and disadvantages of using these systems in microfluidics, drug discovery, and biomedical research.
Table 1. Advantages of microfluidic systems in pharmaceutical and biomedical advancements
Miniaturization & Efficiency | Perform laboratory functions in a microscale environment |
Capacity for miniaturization and parallelization for high-throughput screening | |
Achieve significant reduction in sample and reagent consumption | |
Precision & Control | High surface-area-to-volume ratio enables precise environmental control |
Enhanced analytical sensitivity through microscale reactions | |
Tailored platform designs enable automation and standardization | |
Physiological Relevance | Incorporate complex cell culture techniques with mechanical and biochemical cues |
Improved physiological relevance compared to traditional culture methods |
Together, these capabilities accelerate preclinical workflows and shorten overall timelines for drug discovery and biomedical research. However, despite their advantages, microfluidic platforms still face significant engineering, operational, and translational challenges that hinder broader adoption in pharmaceutical and biomedical research.
Table 2. Limitations of microfluidic systems in pharmaceutical and biomedical advancements
Technical & Manufacturing Challenges | Complex fabrication and integration processes require specialized expertise |
Difficult to scale up for large-scale production and routine implementation | |
Small channels and intricate fluidic networks are highly susceptible to air bubble formation | |
Material properties can limit environmental control and reproducibility | |
Operational & Integration Barriers | Highly specialized designs limit flexibility for broader research applications |
Reliance on proprietary components and consumables complicates integration into conventional laboratory workflows | |
Challenges in adapting platforms for point-of-care applications | |
Regulatory & Standardization Issues | Absence of standardized regulatory guidelines complicates validation and approval processes |
Microfluidic technologies thus offer a powerful bridge between traditional cell culture and clinical research, but achieving full industrial and clinical integration will require addressing current engineering, material, and regulatory barriers that restrict their widespread adoption.
Future directions
Microfluidic technologies in cell culture have rapidly evolved from small-scale experimental tools to viable candidates for industrial and clinical use. Advances in materials, fabrication, and bioengineering have facilitated the transition from proof-of-concept devices to scalable, integrated systems. The growing demand for automation and “hands-off” workflows has driven the creation of user-friendly devices suitable for point-of-care applications. Meanwhile, advances in AI and machine learning are now bridging the gap between high-throughput experimentation and data analysis.
Recent innovations, particularly in 3D printing and microfabrication, have simplified design and production, turning microfluidic OoC systems from specialist-only research methods into accessible tools for microfluidics drug discovery and broader biomedical research. New adsorption-resistant, highly biocompatible materials have enhanced reproducibility and microenvironmental control, while modular “plug-and-play” platforms now enable complex cell cultures with minimal user expertise. Miniaturized biosensors and improved material optical transparency enable real-time monitoring of cellular processes, providing deeper insight into cellular activity. Concurrently, advances in cell culture techniques and complex mechanical and biochemical systems have significantly improved the translational relevance of these models to human physiology.
Despite significant progress in microfluidics drug discovery and biomedical research, some challenges remain before microfluidic systems can achieve full clinical and industrial integration. Greater standardization and clear validation pathways are essential. Further incorporation of patient-derived tissues or iPSC-derived models could enhance the value of microfluidic cell culture devices for drug testing and precision medicine. Continued development of automated and modular systems will be key to achieving broad adoption in biomedical, pharmaceutical, and clinical research.
Conclusion – Transformation of microfluidics drug discovery and biomedical research
Microfluidic technologies are reshaping drug discovery and disease modeling by enabling high-throughput, reagent-savings, and highly precise, biologically relevant models. Droplet microfluidics excel in speed, scale, and cost-efficiency, particularly for antibody and compound library screening. OoC platforms provide physiologically relevant models that capture human-specific responses with greater accuracy than traditional cell-culture or animal models. LoC platforms automate multiple laboratory processes in a single device, reducing the need for human intervention and greatly increasing experimental throughput.
Despite these advantages, important challenges remain. Barriers to the full adoption of microfluidic technologies in drug development workflows and disease include integration with existing processes, the lack of standardized regulatory frameworks, and the need for further engineering development to support fully automated, large-scale deployment. Addressing these challenges will be key to translating microfluidics into mainstream drug development.
Overall, the precision, reproducibility, and predictive power of microfluidic systems position them as a cornerstone technology for accelerating and reducing risk in drug discovery pipelines. With continued innovation and regulatory alignment, droplet and OoC platforms are promising tools in the transformation of microfluidics drug discovery and biomedical research.
References
[1] L. J. Marshall, J. Bailey, M. Cassotta, K. Herrmann, and F. Pistollato, “Poor Translatability of Biomedical Research Using Animals – A Narrative Review,” Altern. Lab. Anim. ATLA, vol. 51, no. 2, pp. 102–135, Mar. 2023, doi: 10.1177/02611929231157756.
[2] N. G. Frangogiannis, “Why animal model studies are lost in translation,” J. Cardiovasc. Aging, vol. 2, no. 2, p. 22, Apr. 2022, doi: 10.20517/jca.2022.10.
[3] “Discovery Phase in Drug Development,” BioAgilytix. Accessed: Oct. 17, 2025. [Online]. Available: https://www.bioagilytix.com/solutions/phases/discovery-phase-drug-development/
[4] H. Matthews, J. Hanison, and N. Nirmalan, “‘Omics’-Informed Drug and Biomarker Discovery: Opportunities, Challenges and Future Perspectives,” Proteomes, vol. 4, no. 3, p. 28, Sept. 2016, doi: 10.3390/proteomes4030028.
[5] D. Zhang et al., “Innovative Advances in Droplet Microfluidics,” Research, vol. 8, p. 0856, doi: 10.34133/research.0856.
[6] “Droplet microfluidics for single cell analysis: The next generation of capability.” Accessed: Oct. 21, 2025. [Online]. Available: https://www.selectscience.net/article/droplet-microfluidics-for-single-cell-analysis-the-next-generation-of-cabability
[7] S. Deng et al., “Organ-on-a-chip meets artificial intelligence in drug evaluation,” Theranostics, vol. 13, no. 13, pp. 4526–4558, Aug. 2023, doi: 10.7150/thno.87266.
[8] J. D. Caplin, N. G. Granados, M. R. James, R. Montazami, and N. Hashemi, “Microfluidic Organ‐on‐a‐Chip Technology for Advancement of Drug Development and Toxicology,” Adv. Healthc. Mater., vol. 4, no. 10, pp. 1426–1450, July 2015, doi: 10.1002/adhm.201500040.
[9] N. Farhang Doost and S. K. Srivastava, “A Comprehensive Review of Organ-on-a-Chip Technology and Its Applications,” Biosensors, vol. 14, no. 5, p. 225, May 2024, doi: 10.3390/bios14050225.
[10] C. M. Ardila, “Advancing healthcare through laboratory on a chip technology: Transforming microorganism identification and diagnostics,” World J. Clin. Cases, vol. 13, no. 3, p. 97737, Jan. 2025, doi: 10.12998/wjcc.v13.i3.97737.
[11] J. Castillo-León and W. E. Svendsen, Eds., Lab-on-a-Chip Devices and Micro-Total Analysis Systems: A Practical Guide. Cham: Springer International Publishing, 2015. doi: 10.1007/978-3-319-08687-3.
[12] H. Azizgolshani et al., “High-throughput organ-on-chip platform with integrated programmable fluid flow and real-time sensing for complex tissue models in drug development workflows,” Lab. Chip, vol. 21, no. 8, pp. 1454–1474, Apr. 2021, doi: 10.1039/D1LC00067E.
[13] N. Shembekar, C. Chaipan, R. Utharala, and C. A. Merten, “Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics,” Lab. Chip, vol. 16, no. 8, pp. 1314–1331, 2016, doi: 10.1039/C6LC00249H.
[14] L. Ming et al., “Microfluidic-based screening of resveratrol and drug-loading PLA/Gelatine nano-scaffold for the repair of cartilage defect,” Artif. Cells Nanomedicine Biotechnol., vol. 46, no. sup1, pp. 336–346, Oct. 2018, doi: 10.1080/21691401.2017.1423498.
[15] Herland, A., Maoz, B. M., Das, D., Somayaji, M. R., Prantil-Baun, R., Novak, R., … & Ingber, D. E. (2020). Quantitative prediction of human drug pharmacokinetic responses using multiple vascularized organ chips coupled by fluid transfer. Nature biomedical engineering, 4(4), 421.
[16] Z. A. Li and R. S. Tuan, “Towards establishing human body-on-a-chip systems,” Stem Cell Res. Ther., vol. 13, p. 431, Aug. 2022, doi: 10.1186/s13287-022-03130-5.
[17] Maoz, B. M., Herland, A., FitzGerald, E. A., Grevesse, T., Vidoudez, C., Pacheco, A. R., … & Parker, K. K. (2018). A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nature biotechnology, 36(9), 865-874.
[18] T. Shroff et al., “Studying metabolism with multi-organ chips: new tools for disease modelling, pharmacokinetics and pharmacodynamics,” Open Biol., vol. 12, no. 3, p. 210333, doi: 10.1098/rsob.210333.
[19] Rauti, R., Navok, S., Biran, D., Tadmor, K., Leichtmann-Bardoogo, Y., Ron, E. Z., & Maoz, B. M. (2023). Insight on bacterial newborn meningitis using a neurovascular-unit-on-a-chip. Microbiology Spectrum, 11(3), e01233-23.
[20] J. Zhai et al., “Drug screening on digital microfluidics for cancer precision medicine,” Nat. Commun., vol. 15, no. 1, p. 4363, May 2024, doi: 10.1038/s41467-024-48616-3.
[21] L. Moll, J. Pihl, M. Karlsson, P. Karila, and C. I. Svensson, “A Microfluidic High-Capacity Screening Platform for Neurological Disorders,” ACS Chem. Neurosci., vol. 15, no. 2, pp. 236–244, Dec. 2023, doi: 10.1021/acschemneuro.3c00409.



Funding and Support
This review was written by Emma Glickman.
Published in October 2025.
Contact: Partnership[at]microfluidic.fr

Check the other Reviews
FAQ - Microfluidics drug discovery and biomedical research
Exactly what topics appear in the review?
A look unfolds – small but sharp – into microfluidic tools like droplet handling, organ-on-a-chip, and lab-on-a-chip. These setups shift ground in early research and tests before clinical trials begin. What stands out? Better prediction, faster results, less fluid waste. Progress is clearly evident in drug candidate checks, live-cell measurements, absorption and effect tracking, and models mimicking human illness.
What drives scholars to shift from conventional frameworks?
Translation falters too often. Most drugs showing promise in animal trials – about 90 % – do not work in people. This high failure rate stems from limited biological accuracy and the sluggish pace of traditional lab cultures. With microfluidic technology, conditions inside test environments become more precise. At the same time, processing speed increases dramatically.
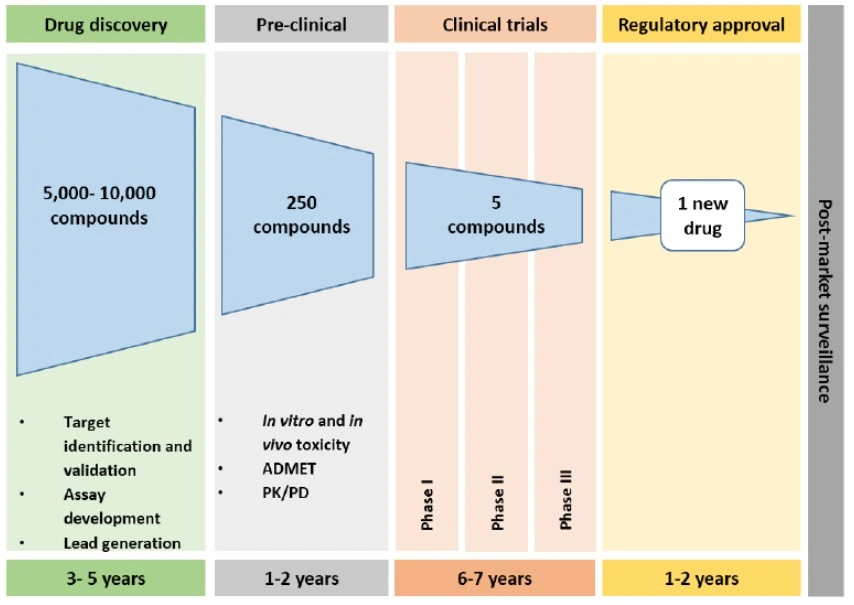
What are the headline numbers worth remembering?
Despite progress, getting a single drug to market often takes about 15 years, and only 1 in 5,000 early compounds succeed. In contrast, advanced lab-on-a-chip systems now capture more than 10,000 images in 60 minutes, while maintaining RNA integrity for later gene analysis. These realities – one slow, one fast – are why automated microfluidics have become essential. The gap between sluggish development and rapid measurement drives innovation.
Droplet microfluidics: what is the specific advantage for screening?
A single drop becomes a tiny enclosed space where reactions happen, consistent in size down to billionths of a liter. Because they are identical, differences in results shrink; their small scale uses far less material; running many at once packs vast numbers into the area of a standard lab plate. When finding antibodies, sorting artificial gene sets, or testing drug mixtures, these drops lower the expense per test while boosting data reliability – the kind of efficiency needed before trials in living organisms.
Organ-on-chip systems: are they really more predictive?
It might not fix everything, yet the case behind it holds weight because they capture physical forces like shear and stretch, along with mixed cell types and three-dimensional structures absent in flat cultures. While single-organ chips target damage specific to individual organs, connected setups mimic absorption, distribution, metabolism, and excretion processes directly on the lab platform. As a result, problems show up sooner, and reactions unique to humans become visible much earlier.
And lab-on-chip – how is it different from OoC?
Automation occurs through the integration of preparation, reaction steps, fluid handling, controlled gradients, and analysis – all within one platform. Programmed liquid paths combine with quick heat shifts and minimal spread between points. This approach enables full assay execution without manual intervention, delivering consistent results across multiple tests at once. On-device measurements such as barrier resistance, gas levels, and molecular flow fit seamlessly into the process. Imaging with rich data output aligns well with these systems.
What new applications feel “ready now” rather than speculative?
• Functional readouts at scale: barrier integrity, metabolism, transport, and morphology – run in parallel with automated imaging and on-chip electrodes.
• High-throughput compound triage: droplet screens and multiplexed OoCs to rank candidates before animal work.
• PK/PD and toxicity workflows: linked tissue models approximating ADME behavior to catch liabilities earlier.
What constraints might appear less obvious?
Hidden expenses often emerge where least expected.
Though standardization efforts continue to evolve, regulatory approval may take time. From an engineering standpoint, challenges include how materials interact with fluids, handling air bubbles, maintaining sterile conditions over months, and connecting microchips to conventional laboratory equipment. When it comes to daily operation, staff must understand fluid behavior – such as pressure regulation, sensor response, and system tuning – alongside building reliable infrastructure to handle vast streams of imaging output and continuous signal recordings.
What might work for a consortium aiming to apply these platforms in Horizon Europe?
A practical approach would start there.
A single tool linked clearly to your project’s performance goals comes first – say, sorting leads by droplet method or testing liver-lung reactions together across organs. Next, assign funds without guesswork: cover liquid movement tech like regulators, drivers, or pressurized setups, then monitoring tools and information storage layers. Bring in a specialist firm skilled in tiny structure production to shorten test-and-refine loops. Add an expert able to run equipment daily while supplying tailored microchips under tight deadlines.
Where does the Microfluidics Innovation Center stand in this context?
A small company focused entirely on microfluidics, MIC handles fluid system engineering, chip layouts, the production of tiny structures, sensor integration, and automated lab processes. Within European Union project groups, an effective small business can boost how proposals are rated for execution and practical use; based on what we’ve seen, MICs usually double the likelihood of getting funded compared to standard rates. On regular timelines, support includes guiding teams through choosing suitable formats – like droplets, organ-on-chip, or labs-on-chips – structuring test setups, producing hardware, and then refining repeatedly until results satisfy defined standards. Rapid prototyping is part of this, too, ensuring high flexibility without a long-term commitment to a single version.
What does a “good” microfluidic assay look like in practice?
Starting with the biology, it traces back through flow conditions, shear forces, dwell times, alongside sensing tools. Where needed, material choices or coatings handle molecule sticking. Each pump and detector comes with full calibration records. Should the research direction shift, there is a built-in capacity to retrieve RNA or protein. Trustworthy outputs emerge – repetition, reference samples, clear quality checks – ready for analysis.
How does the immediate future look?
One likely path forward involves tighter integration of artificial intelligence into experimental workflows. Improved optical access and integrated biological sensors may soon become routine. Modular systems, designed for real-world lab upkeep, could gain traction. What might change things in the coming years is agreement on common standards. Without consistent testing frameworks, cross-lab comparisons risk distortion. Shared evaluation methods would help results move seamlessly between environments.