DNA repair with molecular machines: DNARepairMan
Author
Kos Breiev, PhD
Publication Date
March 16, 2017
Status
Keywords
DNA Repair
genome stability
molecular machines
tumor suppression
mechanistic biophysics
quantitative molecular biophysics
structural biology
Your microfluidic SME partner for Horizon Europe
We take care of microfluidic engineering, work on valorization and optimize the proposal with you
Twelve European organizations from academia and the private sector, from the Netherlands, Deutschland, France, the UK, and Poland, will constitute a research network to study DNA repair.
The main idea behind the DNARepairMan Project is to understand some of the critical human DNA reparation pathways.
Molecular machines for DNA repair pathways: introduction
We can count 1 million molecular lesions per cell daily in our bodies. These lesions lead to DNA damage, which can cause unregulated cell division and, consequently, tumors.
To fight against the damage, DNA repair is constantly active, and a profoundly mechanistic understanding of its pathways is now fundamental. Indeed, the ability of our cells to repair is vital for the integrity of the genome.
The relevance of this question has been recognized thanks to the 2015 Nobel Chemistry Prize, granted to three researchers focusing on DNA repair. Thus, the main goal of the DNARepairMan project is to perpetuate the virtuous cycle between new technologies, new questions, and new insights.
Molecular machines for DNA repair pathways: project description
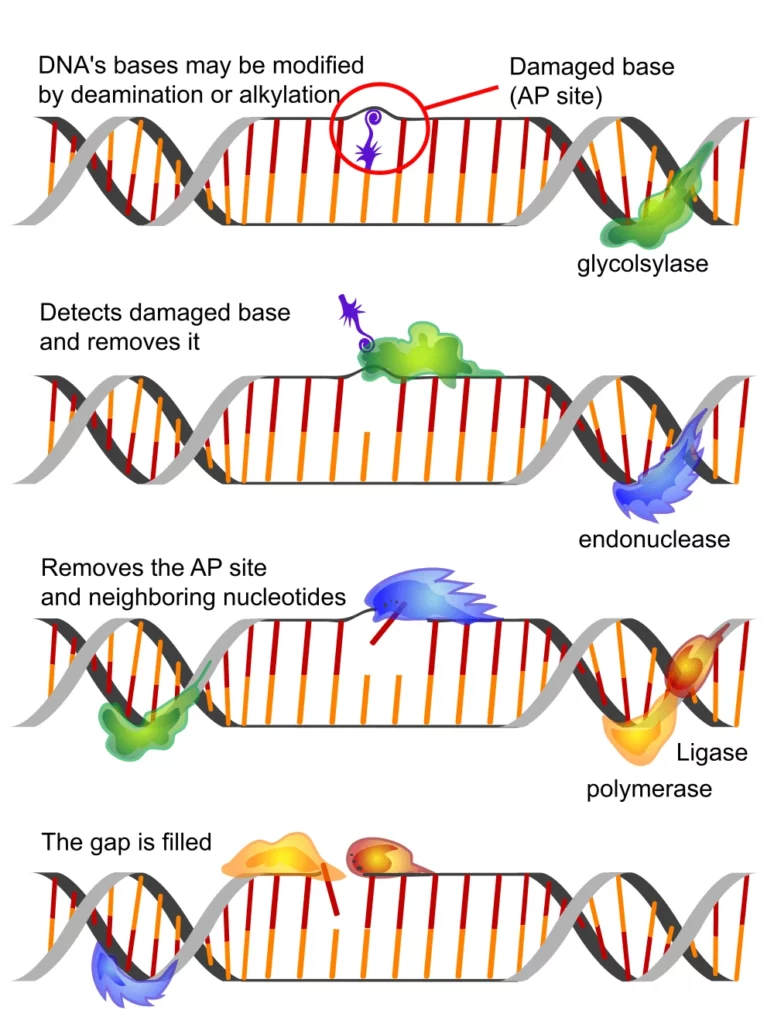
The DNARepairMan project gathers scientists from three areas of research: biologists, chemists, and physicists.
It aims to answer a fundamental research question: what are the switches and motors’ statistical properties and molecular mechanisms involved in two critical DNA repair pathways?
Thus, the objectives of this consortium will be to:
- Characterize the mechanism of lesion formation.
- Determine the structure of the helicase recruitment complexes.
- Characterize the catalytic properties of the unwinding complexes.
- Understand the regulation of their activity and establish the link between DNA repair and replication.
To address this challenge, a DNA-Paint microfluidic platform was developed and implemented at LMB and CNRS, to multiplex single-molecule light microscopy experiments.
Check our application note about microfluidic colocalization setup for DNA analysis.
This project has received funding from the European Union’s Horizon research and innovation program under the Marie Sklodowska-Curie grant agreement No 722433 (DNARepairMan project).
Researcher

Kos Breiev
PhD candidate
- MSc in chemistry and water treatment from Kiev University
- Applied Physics and software development (Java, Labview) at the University of Innsbruck, Austria
Areas of expertise:
Organic chemistry, applied physics, plasma, software, microfluidics



Check our Projects
FAQ – DNA repair with molecular machines: DNARepairMan
What is DNArepairMAN?
It is an R&D project to create and test so-called molecular machines that can detect DNA damage and support or direct endogenous repair pathways. Consider programmable nanoscale assistants that dock onto DNA damage, prefer high-fidelity repair products, and minimize error-prone repair products that propagate mutations.
What is the rationale of repairing the target DNA and not the damage?
Due to the constant nature of damage (oxidation, alkylation, UV lesions, double-strand breaks, etc.), cells already consume much energy to repair it. Instead of introducing brute-force chemistry, it is possible to enhance the quality of the genome by adding a repair toolkit (BER, NER, HR, NHEJ) by nudging it, rather than adding brute force.
What is the name of a so-called molecular machine in this case?
Multi-domain construct, in which three functions are coupled together: (i) lesion recognition (sequence/structure-specific binding), (ii) local actuation (recruitment of a repair enzyme, steric masking of error-prone factors, or catalytic hand-off), and (iii) readout (fluorescent or electrochemical signal to confirm engagement). The forms range from engineered proteins and peptide scaffolds to polymers, polymer-protein hybrids, and DNA-origami.
What were the scientific use-cases that were of interest to the team?
Three recurring ones:
- High-fidelity repair bias in model cell lines in which the double-strand breaks are likely to be repaired incorrectly.
- Intervention-based damage mapping, i.e., sensing 8-oxoG or abasic sites and recruiting a clean repair pathway during the same step.
- Small-scale genotoxicity assay miniaturization, reducing large genotoxicity assays to chip-scale, quantitative outputs.
What is the actual measure of better repair?
The combination of orthogonal readouts: (i) lesion-specific qPCR dropouts, which resolve during repair, (ii) reporter cassettes, which flip only in cases of error-free repair, and (iii) single-cell imaging of g-H2AX foci dissipation. We use the combination of fluorescence time-series and endpoint sequencing on chip to measure the speed and fidelity.
Does the approach focus on an indication or on an entire disease?
Mechanically indifferent, clinically discriminatory. The platform will compare machinery against general damage first and then will further refine to those settings where fidelity is the most important e.g., pre-neoplastic clones, neurodegeneration model with oxidative stress or cell-therapy manufacturing where genomic scars should be minimized.
What was the specific contribution of MIC?
Design-for-experimentation. We delivered:
- Microfluidic assay carriers based on stable and low-autofluorescence materials compatible with live-cell imaging.
- Flow control and automation such that every chip operates with specific residence times and shear rates (0.1-10 Pa) with no drift.
- Measurement, e.g., inline measurements, imaging-friendly windows, and data interfaces to downstream measurements.
Do these machines serve a therapeutic but not a research purpose?
They are research-quality with a translational focus. The near-term value lies in accurate phenotyping of repair, mapping biases, quantifying rescue, and head-to-head comparisons of candidacy. The gating items are in clinical, delivery, immunogenicity, and off-target binding; they are being de-risked with a parallel targeted format.
To what extent are the chip-based assays inter-site reproducible?
By compressing channel cross-sections, flow programs, and gasket compression, we can push channel variability to single-digit percent with standard readouts. The protocol template also includes calibration beads and internal standards to ensure reproducibility of curves within confidence limits acceptable to a remote site.
Does it deal with primary or patient-derived cells?
Yes, with modified shear and ECM coating. We have performed primary epithelial and neuronal cultures by reducing wall stress, using oxygen-permeable materials, and adding perfusion breaks to circumvent metabolic fatigue. Some delicate phenotypes are even stabilized in the microfluidic environment.
What is success at the end of the project?
A confirmed microfluidic workflow for a candidate molecular machine: (i) localizes to specific lesions, (ii) demonstrates substantial enhancement in high-fidelity repair compared to controls, and (iii) can be characterized by a second site and can be recapitulated within established limits. In addition, a prototype kit featuring SOPs to allow new partners to run it without re-engineering the system.