Elucidating human enhanceropathies using droplet microfluidics: ENHPATHY
Author
Robert Baber, PhD
Publication Date
October 10, 2019
Status
Keywords
Single-Cell Omics
Lab-on-a-chip
droplets
high-throughput screening
gene regulation
enhanceropathies
single-cell encapsulation
Your microfluidic SME partner for Horizon Europe
We take care of microfluidic engineering, work on valorization and optimize the proposal with you
Elucidating human enhanceropathies using droplet microfluidic: introduction
Enhancers are regulatory DNA sequences that utilize various mechanisms to control gene expression. Enhancer malfunction caused by either genomic or epigenomic alterations can lead to multiple diseases. These are termed enhanceropathies and can range from congenital disorders (e.g., polydactyly) and autoimmune diseases (e.g., diabetes) to cancer.
Considering the increased incidence of some of these diseases in recent years, there is an urgent need for new diagnostic and therapeutic strategies to combat them. Developing effective methods requires a detailed understanding of the molecular basis of human enhanceropathies.
Elucidating human enhanceropathies using droplet microfluidic: project description
Conventional methods to interrogate enhancer function are insensitive to cell-to-cell variations and only reflect population-level views. This is a significant shortcoming, as cellular heterogeneity is inherent to most tissues and cell populations.
To decipher the full molecular complexity of enhanceropathies, it is essential to analyze a large number of cells from complex samples individually. This can be achieved using droplet microfluidics, which enables high-throughput single-cell encapsulation and thus allows the study of enhancer function at single-cell resolution.
This project aims to develop systems based on droplet microfluidics that help other groups within the ENHPATHY consortium study enhancer function at single-cell resolution, thus, shedding light on the molecular mechanisms of enhanceropathies.
We will use the OB1 flow controller and the MFS flow sensor (Elveflow) to obtain systems enabling automated, high-throughput droplet production with precise control over droplet size. Moreover, droplet production on these systems will be highly parallelized, increasing throughput for single-cell encapsulation.
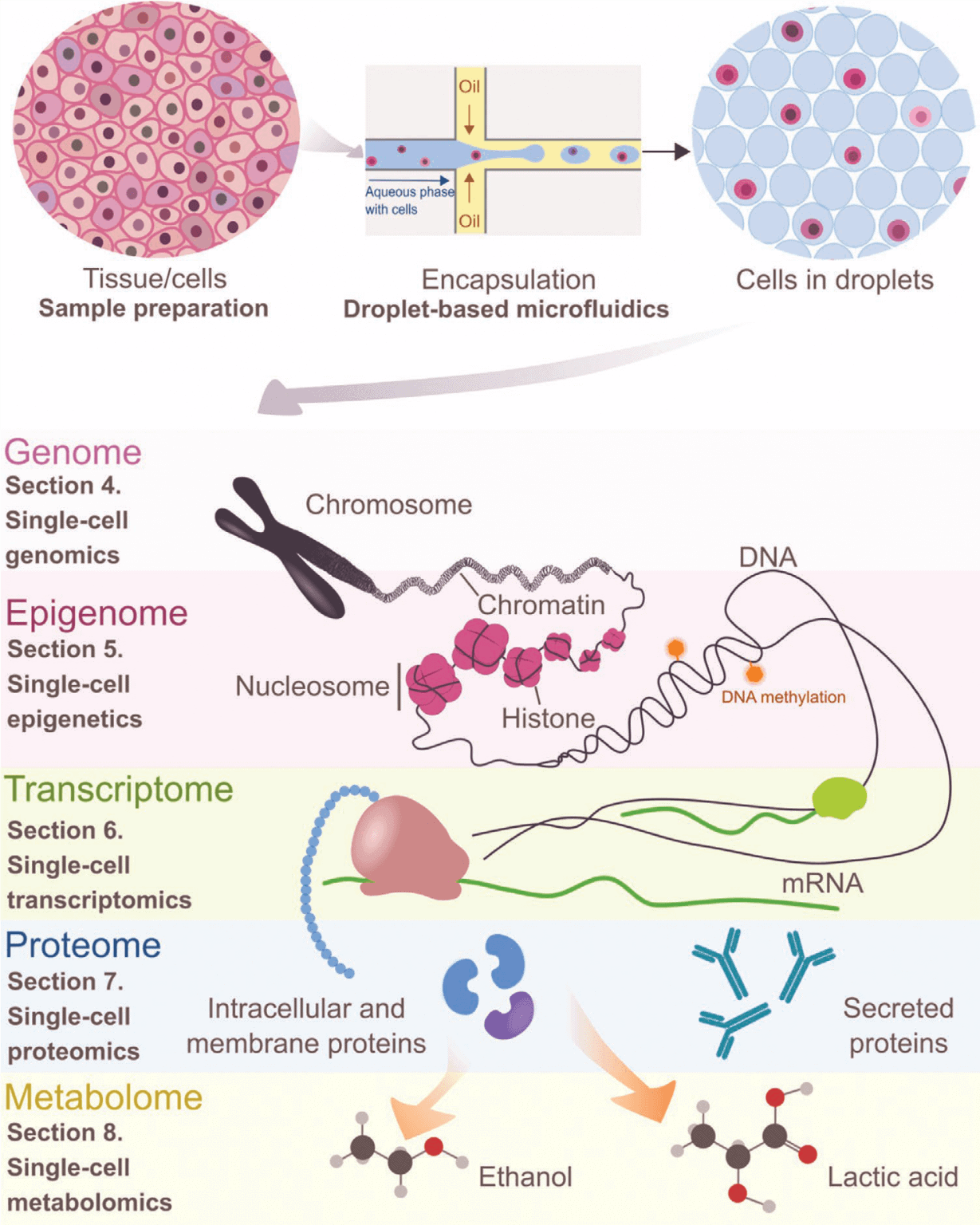
Finally, the commercialization potential of the aforementioned systems will be investigated. Ideally, this project results in a commercially available microfluidic system that can be customized, is easy to use (plug and play), and enhances or simplifies research in single-cell omics, including studying enhanceropathies.
Thus, this would ultimately also facilitate the translation of molecular findings into new diagnostic and therapeutic avenues for patients suffering from diseases such as enhanceropathies.
Results from this project
This project helped develop the inflammatory bowel disease pack and the precision sampling for cell culture.
Funding
The European Union’s Horizon 2020 MSCA-ITN has funded the Enhpathy project under grant agreement No. 860002 (ENHPATHY project).



Researcher

Robert Baber
Marie-Curie PhD candidate Elvesys/KTH
- R&D Engineer in Microfluidic Cartridge Development Team, BluSense Diagnostics (Copenhagen, Denmark)
- M.Sc. Eng. in Physics and Nanotechnology, Technical University of Denmark (Copenhagen, Denmark)
Areas of expertise:
Microfluidics, Micro- & Nanofabrication, Physics, Lab-on-a-Chip, Computer-aided design.
Check our Projects
FAQ – Elucidating human enhanceropathies using droplet microfluidics: ENHPATHY
What is meant by enhanceropathies?
Enhanceropathies are disorders whereby non-coding regulatory DNA particularly enhancers malfunction. The sequence can be mutated itself, the enhancer can be displaced (or, sometimes, by means of structural variants), or the 3D interactions of the protein and the chromatin can be altered. The resultant phenotype comprises mis-timed or minor percentage gene expression with tissue-specific phenotypes: characteristic of limb malformations, heart anatomy flaws, neurodevelopmental syndromes, and cancer subsets.
So why is droplet microfluidics a good application in enhancer biology?
Droplets of water in oil enable you to run tens of thousands of single-cell or single-construct experiments simultaneously, each in a picoliter-scale reactor, thereby conserving transcriptional noise and burst dynamics. On typical platforms, the droplets per chip are 10^5–10^6, volumes are approximately 10–100 pL, and there is approximately a 50× reduction in reagent costs compared to plate-based screens.
What experiments can be performed by droplets?
Three broad families:
• Massively parallel reporter assays (MPRA): barcoded collection of enhancers entrap cells or cell-free systems in order to measure distributions of activity and not just means.
• Perturb-seq in droplets: CRISPRi/a plus scRNA-seq readouts.
• Biochemical reconstitution: Phase separation/nucleosome logic, minimalist, cell-free mixes to test TF-enhancer-cofactor logic.
What are the scale characteristics of readout?
Two routes are standard. (i) Optical: optical reporters of any of the previously discussed methods, e.g., the simplest: fluorescence of dropped material, on-chip photometers, etc. Droplets could be sorted at 500-2,000 Hz by intensity (or ratiometric logic). There are two other methods, staples of microfluidics: (ii) Sequencing droplets that are barcoded through microfluidic co-flow or bead-based barcodes; and (iii) absolute distributions of activity and identities of enhancers-guides. However, in practice, the laboratory may still use both: optical sorting to pre-enrich, and then perform NGS.
What membership of this project did MIC actually engineer?
The droplet-on-demand workflow is enhanced with a regulatory genomics workflow: droplet-based generation with clean droplets (CV in droplet diameter normally <3%), inducers delivered on-demand by merging droplets, and dielectrophoretic sort also allows populations with low expression to be sorted. This was bundled with SOPs for reporter design, oil/surfactant chemistries to enable downstream PCR, and quality-control measures (encapsulation Poisson λ, barcode collision rate, commonly yield of recovery) to allow runs to be comparable across sites.
What are some of the tangible biological, i.e., practical, questions that this can answer?
Examples that we are used to seeing: (i) heightening effects with alleles at heterozygous backgrounds; (ii) shadow enhancers and stress-induced redundancy; (iii) transcriptional bursting, does variation in burst size depend on frequency?; (iv) 3D rewiring, is it possible to boost adoption of enhancers following recreation of a pathogenic structural variant?
What are the performance statistics of a team on its first campaign?
A viable baseline: 150-300,000 droplets/run to be analysed, >90 percent barcode recovery of NGS readouts, purity of optical sorting ≈95 percent at around 1 kHz, and 50 μL of reagent used/condition. With MPRA-style libraries, power calculations indicate that approximately 1,000-5,000 observations per enhancer version detect 10-20% changes in mean activity with power >0.9.
What is the way in which data quality is controlled?
The variables observed are λ (number of cells per droplet), co-encapsulation, rates of sort gates on/off carryover, and pre/post-sort distributional (KL divergence vs. target). At the sequencing end: UMI saturation curve, barcode collision, and accuracy of linkage of dispensing enhancers. Sources that do not pass preset criteria are rerun with modified flow conditions or surfactants; this is baked into the SOP.
Does this pipeline have the capacity to provide clinical or translational projects?
With proper governance, yes. Because diagnostics are closely related (e.g., variant interpretation), we use batch controls, spike-ins (absolute calibration), and pre-registered analysis plans. For therapeutic discovery, droplet assays are ideal filters; evidence shows they selectively identify enhancer targets whose modulation restores distributions of expression toward healthy baselines.
What would cooperation with MIC be in Horizon Europe?
We typically assume the responsibility of microfluidic format, high-throughput prototyping, assay automation, and dissemination/exploitation packages. The presence of an effective SME can be expected to bolster performance; in multiple European consortia, success probability increases by about 2× relative to baseline with the inclusion of MIC. Other milestones such as early prototypes and risk-reduction are also provided.
What would you like us to do to commence?
Detailed dossier: disease model candidates; list all enhancer candidates or variant classes; list all possible readouts (optical vs. NGS), cell type(s), and biosafety constraints, up to throughput expectations. We simply need a set of barcodes mapped to the droplet generation and recovery steps, assuming you already have reporter libraries or CRISPR designs of interest.