Droplet microfluidics in action
From innovative applications to core challenges
Author
Celeste Chidiac, PhD
Publication Date
June 26, 2025
Keywords
Droplet Microfluidics
Single-cell analysis
Drug screening
DNA construction
Tissue engineering

Need advice for your droplet microfluidic system?
Your microfluidic SME partner for Horizon Europe
We take care of microfluidic engineering, work on valorization and optimize the proposal with you
Introduction to droplet microfluidics
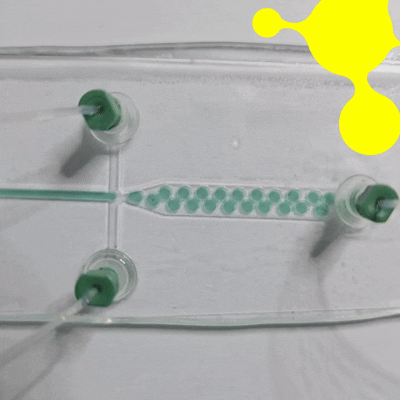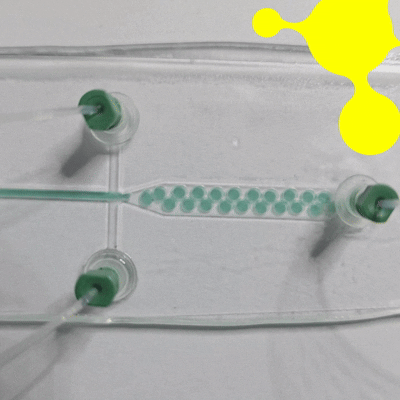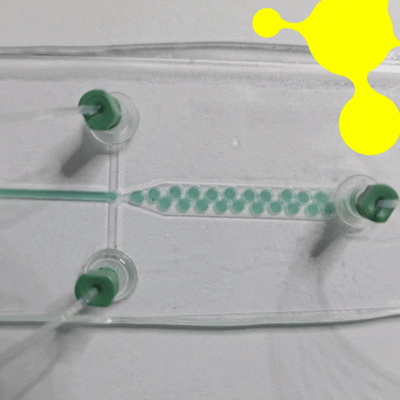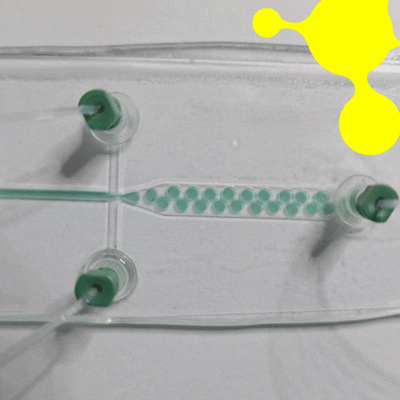
Droplet microfluidics is a technology for studying the generation, manipulation, and application of femtoliter to nanoliter drops encapsulated into monodisperse droplets (Figure 1). In practice, this means that preparation, analysis, and detection can all take place within a single droplet. Compared with bulk experiments, reactions are faster, less reagent is wasted, and many tests can be run at the same time. The method is also more sensitive and gives researchers tighter control at each step [1, 2].
Droplet microfluidics advantages
Traditional life science methods are often costly and depend on complex cell labeling. Sample preparation can take time, which may compromise cell health and increase the risk of contamination. In addition, these approaches are limited in their detection ability [3]. On the other side, miniaturized plates can only support static cell cultures, unable to replicate the dynamic extracellular microenvironment necessary for accurate biological studies.
Microfluidics is quickly becoming a go-to tool in life sciences for good reason. It uses small amounts of material, delivers highly sensitive results, and fits easily into larger lab systems. It can be used to perform 3D cell culturing through continuous infusion, simulating the microenvironment associated with cell physiology in vivo [4].
Microwell-based methods are straightforward and easy to set up, which makes them suitable for creating neat arrays of single cells. However, it can be difficult to deliver liquids on the spot because the reagents have to be well spread across the whole array. That’s a problem if you’re trying to track fast changes in cell behavior.
That’s where droplet microfluidics shines. It’s fast, reliable, and doesn’t waste precious reagents. More importantly, it provides researchers with tight control over experiments and is especially useful when studying how cells react in real-time, making it a smart upgrade for many types of cell research [4] (Figure 2).
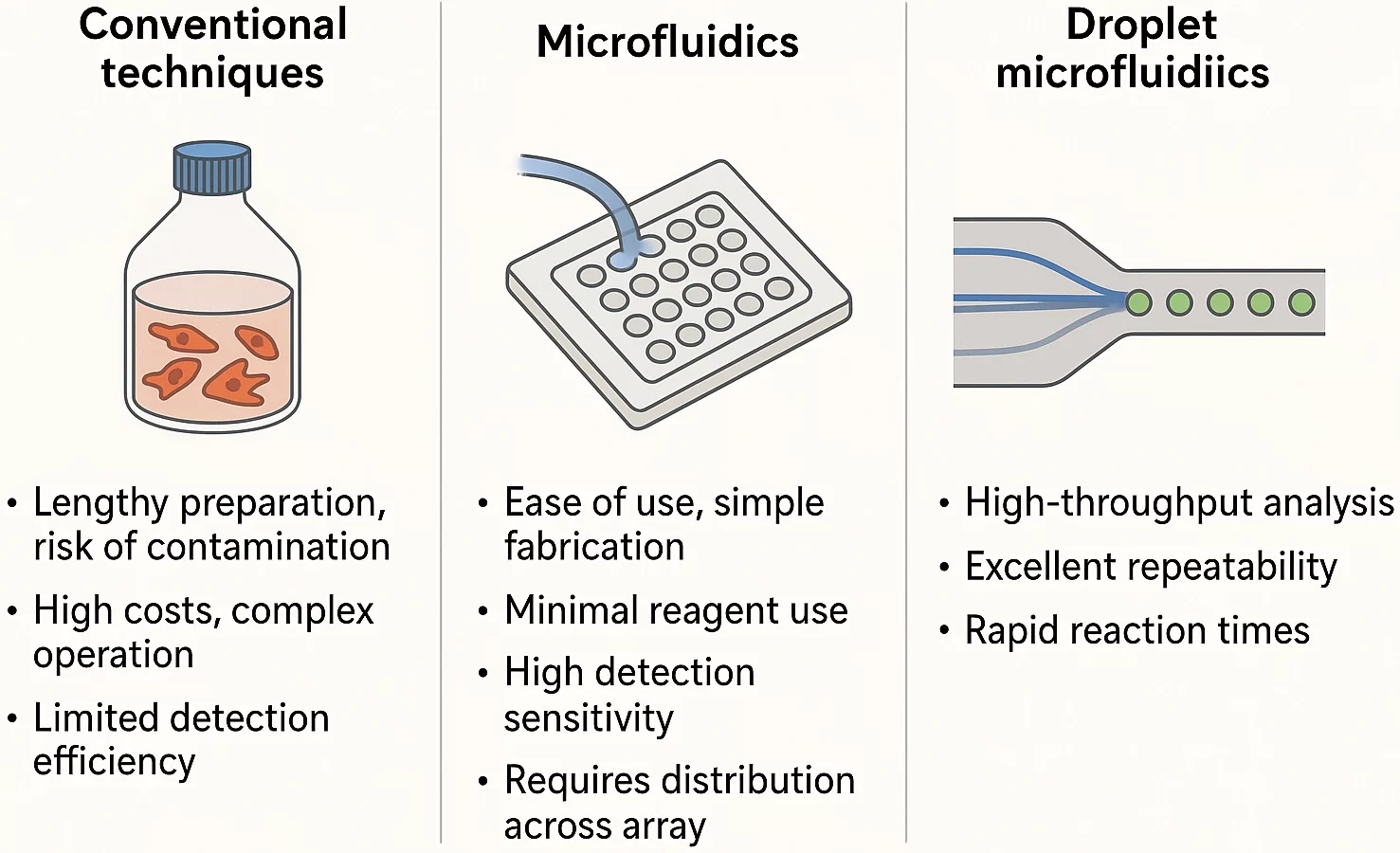
Droplet microfluidics applications
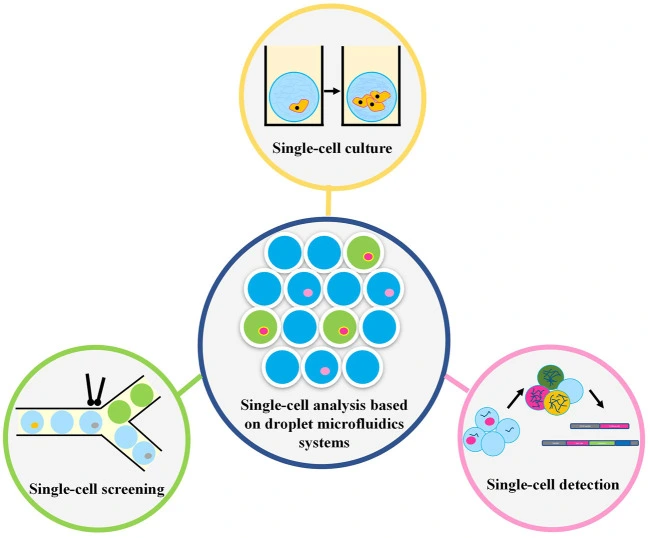
Cell sorting & single-cell analysis
Fluorescence-activated cell sorting (FACS) is widely used for separating cell types, but it mostly works on bulk populations. That makes it hard to study individual cells. It also relies on fluorescent markers, which often demand preparation that can be harsh on the cells [5].
With droplet microfluidics, researchers can study both what’s happening inside the cell (Figure 3) and also at the molecules it releases, without the need for aggressive treatments [1].
Recently, scientists have begun combining droplet microfluidics with traditional FACS, creating hybrid systems that blend FACS’s speed with the precision of single-cell droplet analysis. The result: much higher resolution and control.
For example, Lan and colleagues used droplet microfluidics combined with permeable microgels to sequence the genomes of 50,000 single cells. Their setup enabled them to gently break open the cells and isolate their DNA while preserving its integrity [6].
Rivello and colleagues analyzed the metabolism of individual circulating stromal cells from prostate cancer patients using droplet microfluidics. By measuring pH changes and sequencing mRNA at the single-cell level, they were able to detect highly active cells, highlighting the technique’s strong potential for cancer diagnostics [7].
Drug screening
In microfluidic single-cell drug screening, selected compounds are added to isolated individual cells, not to large cell populations, as is typically done in flow cytometry [8]. This approach eliminates cell-cell interactions and enables the isolation of drug-resistant cells, based on their variable drug sensitivity [4].
For example, Sarkar et al. used a droplet microfluidics setup to study how breast cancer cells absorb and react to doxorubicin. They found that sensitive cells absorbed more of the drug and were more likely to die, often showing bursts of uptake. In contrast, drug-resistant cells showed lower uptake and retention [9].
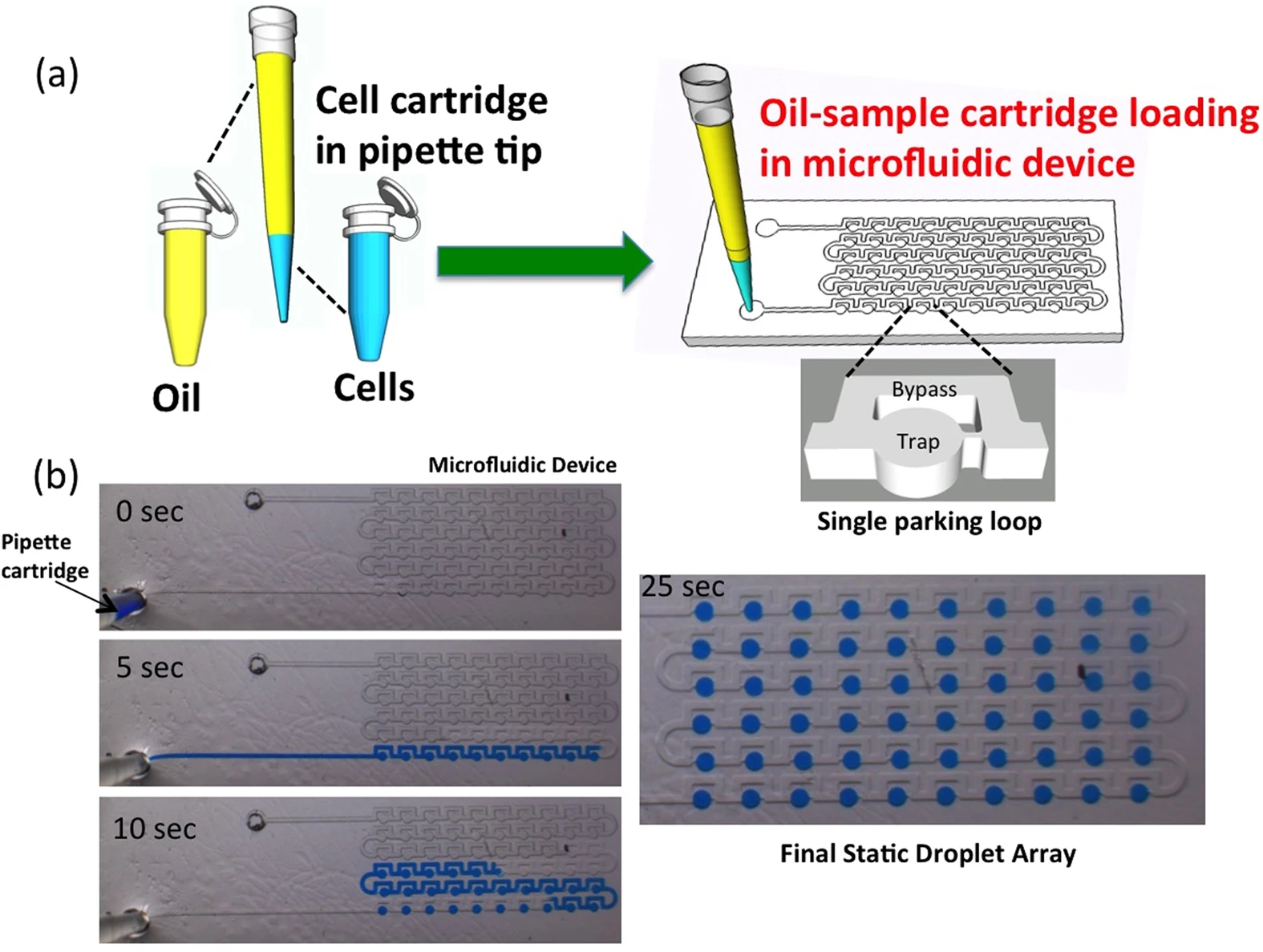
Building on this idea, Bithi et al. developed a simple, pipette-based method to isolate cells and create droplet arrays for analyzing tiny tumor samples (Figure 4). They found that while drug uptake varied among individual cells, cell death only happened once a critical intracellular concentration was reached [10].
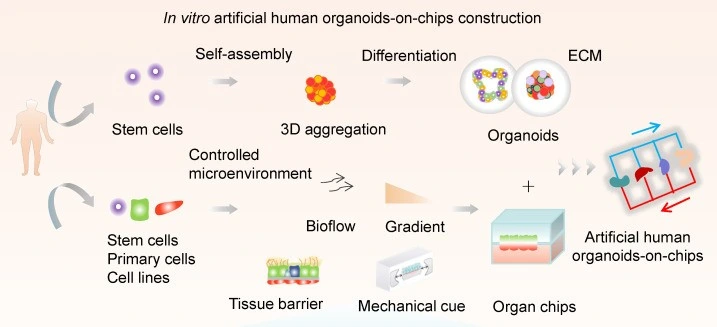
Tissue engineering
By creating 3D structures like tumor spheroids, stem cell spheroids, and organoids (Figure 5), researchers can closely replicate cell behavior, interactions, and differentiation in real human tissues. When combined with cancer-associated fibroblasts (CAFs), these cells provide new insights into tumor resistance to drugs, and allow for the study of tumor stress behaviors under hypoxic conditions [11].
Pushing things even further, researchers are now turning to “organoid-on-a-chip” systems for greater control over nutrient flow and mechanical forces. A team led by Wang developed a one-step method to grow uniform liver organoids from human-induced pluripotent stem cells (hiPSCs) using double emulsion and microfluidic valves [12].
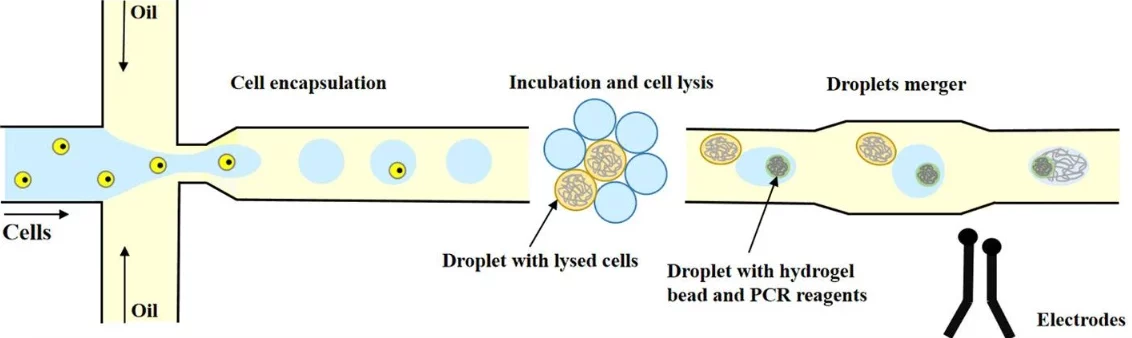
DNA construction in droplets
Microarrays were considered a great advancement in reducing costs and minimizing reaction volumes; however, microfluidics introduced a new level of precision, enabling the individual manipulation of reaction chambers (Figure 6). Digital microfluidic platforms can now automate entire DNA workflows, like constructing gene libraries or assembling large DNA fragments [14]. Yehezkel and colleagues were able to build double-stranded DNA from 160-base-pair pieces inside nanoliter droplets [15].
Coelho and colleagues created a hybrid system that combines digital and droplet microfluidics. First, the digital part carefully mixes the reagents, and then those mixtures are sent into the droplet module, which produces stable, nanoliter-sized droplets. While traditional droplet devices can create a wider variety of droplet sizes and speeds, this hybrid setup provides more consistent and controlled droplets, without slowing down the process [16].
These systems aren’t just for assembling DNA. Shih and his team created a chip that could mix DNA and enzymes, incubate them in winding microchannels, and even deliver electrical pulses to insert the DNA into cells automatically. Meanwhile, Ma and colleagues were able to detect SARS-CoV-2 with high sensitivity and accuracy using a chip that automates nucleic acid extraction and digital droplet PCR [17].
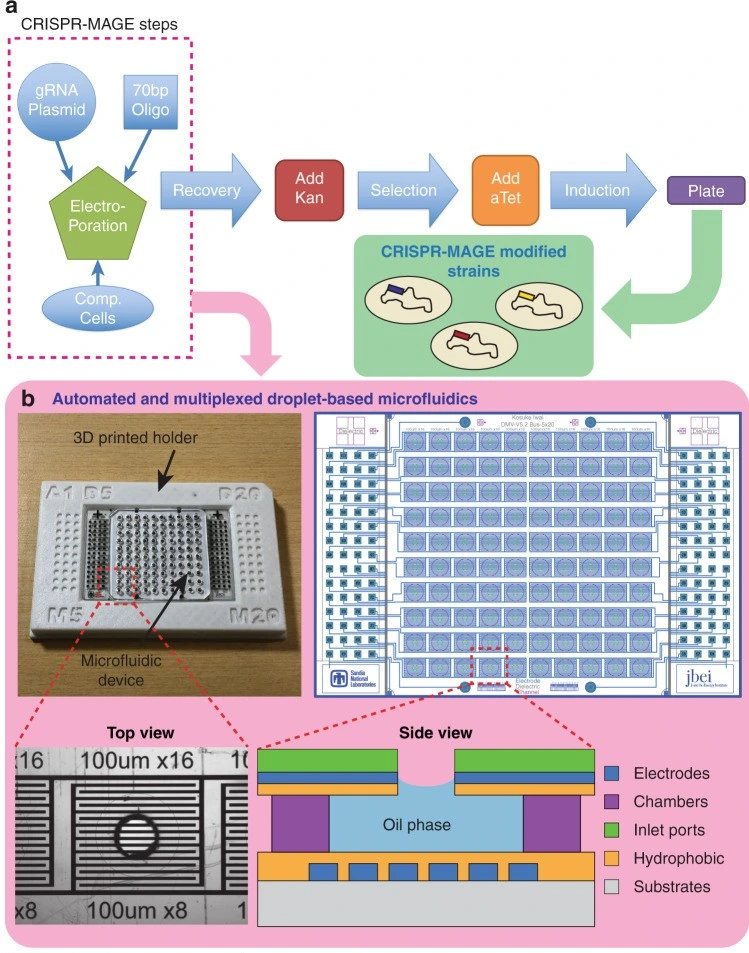
CRISPR-powered microfluidics
Microfluidics is proving to be a game-changer for CRISPR-based gene editing, enabling researchers to automate and miniaturize crucial steps, such as RNA delivery, transfection, and clone selection. This integration leads to faster workflows, reduced reagent use, and more reproducible results – key advantages for scaling up genome engineering [19].
Iwai et al. developed a chip that combines gene editing and testing. It uses a grid of 100 electrodes to move droplets around and zap cells with electric pulses. When they tested it on E. coli bacteria, they were able to enhance metabolite production [20] (Figure 7).
Another team, led by Chen, combined CRISPR with magnetic particles and microfluidics to develop a small device that detects SARS-CoV-2 in under 30 minutes using just 100 µL of sample [21]. It showed high sensitivity, highlighting the potential of CRISPR-microfluidic platforms for rapid, point-of-care diagnostics.
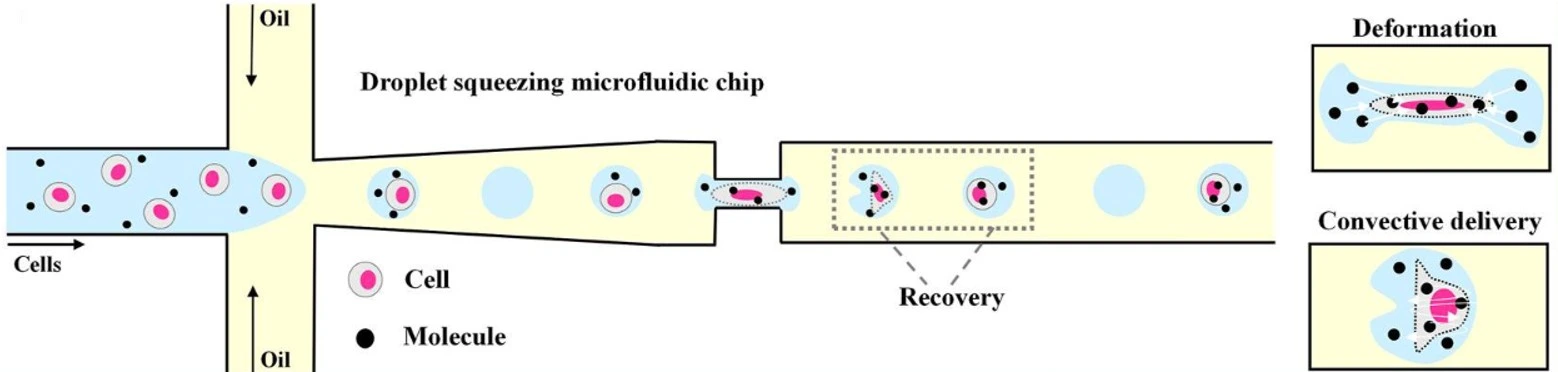
Immunotherapy research
Droplet microfluidics enables highly sensitive functional assays at the single-cell level (Figure 8). This approach helps identify rare immune cells and select the best candidates for cell-based therapies.
One innovative technique consists of anchoring cholesterol-linked antibodies to the cell surface before encapsulation. Cytokines secreted by immune cells are captured directly on the same cell’s surface, eliminating signal mixing between neighboring cells and thereby helping to better distinguish between cell types and activation states [22].
In a similar effort, Yuan and colleagues used droplet microfluidics to co-culture natural killer (NK) cells with their target cells. This setup enabled them to precisely measure the release of interferon-gamma (IFN-γ), a key immune signaling molecule. Their method significantly reduced false results and enabled the sorting of activated cells using standard FACS [23].
Enzyme Kinetics
Recent studies have highlighted the use of droplet microfluidics for enzyme-directed evolution, the study of novel catalytic activity, and the enhancement of protease and peptidase properties (Figure 9).
Okal and colleagues used a high-throughput droplet-based platform to screen libraries of Angiotensin-converting enzyme 2 (ACE2) variants directly on their peptide. The authors were able to highlight beneficial mutations, such as K187T, that significantly enhanced ACE2’s catalytic activity. Their approach demonstrates how microfluidics can facilitate the precise and scalable development of therapeutic enzymes [25].
Schnettler and co-workers recently showed an advancement in the use of droplet microfluidics for enzyme evolution. They took a metal-free α/β-hydrolase and, through droplet screening, turned it into a phosphotriesterase. The reaction they achieved was about a billion times faster than the uncatalyzed version, which made it possible to quickly identify the most active variants [26].
Challenges and future directions
Droplet microfluidics encounters some challenges that hinder its wider adoption (Figure 10).
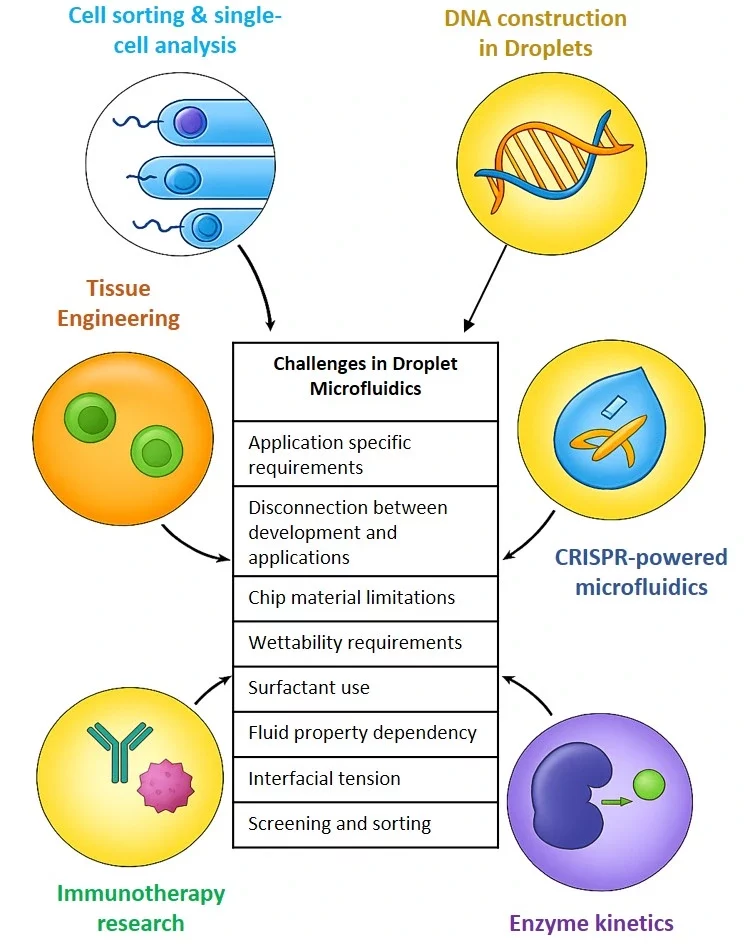
- Application-specific requirements: Each application has very specific requirements for droplet functions, e.g., single-cell analysis needs tiny droplets, whereas tissue engineering requires much larger ones. Technologies for one application can’t simply be scaled or modified for another.
- Disconnection between developers and users: Engineers creating microfluidic systems may not fully understand the daily needs of researchers, while users often find the systems too technical or rigid. Even a small change in an experiment might require a complete redesign of the chip, something most users can not handle.
- Chip material limitations: Choosing the right material can be tricky. Glass chips offer good surface modification options but are expensive (~$500/chip). On the other hand, PDMS chips are cheap and optically transparent, but they are mechanically soft and prone to swelling in oil, which reduces accuracy and compatibility with certain chemicals.
- Wettability requirements: Droplet stability relies on surfactants and surface treatments, which need to be fine-tuned for each experiment.
- Fluid property: Droplet microfluidics is mostly based on Newtonian fluids, which don’t translate well to real biological samples like blood.
- Interfacial tension: Interfacial tension is often considered as constant, which isn’t valid for dynamic or temperature-varying systems (e.g., PCR).
- Screening and sorting: Among droplet manipulations, screening and sorting are the most challenging, as only a small fraction of droplets among millions encapsulate the desired cells [28].
Conclusion - What’s next for droplet microfluidics?
Droplet microfluidics has enabled new possibilities in biology, chemistry, and diagnostics. Its ability to precisely control tiny liquid amounts makes it ideal for high-throughput experiments, single-cell studies, and enzyme screening, saving time, reducing waste, and facilitating discoveries that would be hard to achieve with traditional methods. But alongside these strengths come real challenges. The technology can be complex to design and use, and experiments often need careful tuning to work properly. Even so, there are opportunities for smarter design, better collaboration, and creative problem-solving. As researchers continue to push the limits of what’s possible and make systems more reliable and user-friendly, droplet microfluidics is likely to become an even more powerful tool across science and medicine.
References
- Amirifar, L., et al., Droplet-based microfluidics in biomedical applications. Biofabrication, 2022. 14(2): p. 022001.
- Li, B., et al., Droplets microfluidics platform-A tool for single cell research. Front Bioeng Biotechnol, 2023. 11: p. 1121870.
- Liu, L., et al., Advances of Single-Cell Protein Analysis. Cells, 2020. 9(5).
- Liu, D., et al., Single-cell droplet microfluidics for biomedical applications. Analyst, 2022. 147(11): p. 2294-2316.
- Shields, C.W.t., C.D. Reyes, and G.P. López, Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip, 2015. 15(5): p. 1230-49.
- Lan, F., et al., Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol, 2017. 35(7): p. 640-646.
- Rivello, F., et al., Probing single-cell metabolism reveals prognostic value of highly metabolically active circulating stromal cells in prostate cancer. Sci Adv, 2020. 6(40).
- Drescher, H., S. Weiskirchen, and R. Weiskirchen, Flow Cytometry: A Blessing and a Curse. Biomedicines, 2021. 9(11).
- Sarkar, S., et al., Phenotypic drug profiling in droplet microfluidics for better targeting of drug-resistant tumors. Lab on a Chip, 2015. 15(23): p. 4441-4450.
- Bithi, S.S. and S.A. Vanapalli, Microfluidic cell isolation technology for drug testing of single tumor cells and their clusters. Sci Rep, 2017. 7: p. 41707.
- Wang, Y., et al., Recent methods of droplet microfluidics and their applications in spheroids and organoids. Lab on a Chip, 2023. 23(5): p. 1080-1096.
- Wang, Y., et al., One-step synthesis of composite hydrogel capsules to support liver organoid generation from hiPSCs. Biomaterials Science, 2020. 8(19): p. 5476-5488.
- Wang, H., et al., Human organoids-on-chips for biomedical research and applications. Theranostics, 2024. 14(2): p. 788-818.
- Gach, P.C., et al., Droplet microfluidics for synthetic biology. Lab on a Chip, 2017. 17(20): p. 3388-3400.
- Yehezkel, T.B., et al., Synthesis and cell-free cloning of DNA libraries using programmable microfluidics. Nucleic acids research, 2016. 44(4): p. e35-e35.
- Coelho, B.J., et al., Hybrid Digital-Droplet Microfluidic Chip for Applications in Droplet Digital Nucleic Acid Amplification: Design, Fabrication and Characterization. Sensors (Basel), 2023. 23(10).
- Ma, C., et al., On-Chip Nucleic Acid Purification Followed by ddPCR for SARS-CoV-2 Detection. Biosensors (Basel), 2023. 13(5).
- Pellegrino, M., et al., High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res, 2018. 28(9): p. 1345-1352.
- Azimzadeh, M., et al., CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications. Chemosensors, 2022. 10(1): p. 3.
- Iwai, K., et al., Scalable and automated CRISPR-based strain engineering using droplet microfluidics. Microsyst Nanoeng, 2022. 8: p. 31.
- Chen, F.-E., et al., Point-of-care CRISPR-Cas-assisted SARS-CoV-2 detection in an automated and portable droplet magnetofluidic device. Biosensors and Bioelectronics, 2021. 190: p. 113390.
- Xu, Y., et al., Single-Cell Secretion Analysis via Microfluidic Cell Membrane Immunosorbent Assay for Immune Profiling. Anal Chem, 2024. 96(1): p. 49-58.
- Yuan, Y., et al., Droplet encapsulation improves accuracy of immune cell cytokine capture assays. Lab on a Chip, 2020. 20(8): p. 1513-1520.
- Joo, B., et al., Highly Efficient Transfection of Human Primary T Lymphocytes Using Droplet-Enabled Mechanoporation. ACS Nano, 2021. 15(8): p. 12888-12898.
- Okal, E.F., P.A. Romero, and P. Heinzelman, Droplet microfluidic screening to engineer angiotensin-converting enzyme 2 (ACE2) catalytic activity. Journal of Biological Engineering, 2025. 19(1): p. 12.
- Schnettler, J.D., et al., Ultrahigh-Throughput Directed Evolution of a Metal-Free α/β-Hydrolase with a Cys-His-Asp Triad into an Efficient Phosphotriesterase. Journal of the American Chemical Society, 2023. 145(2): p. 1083-1096.
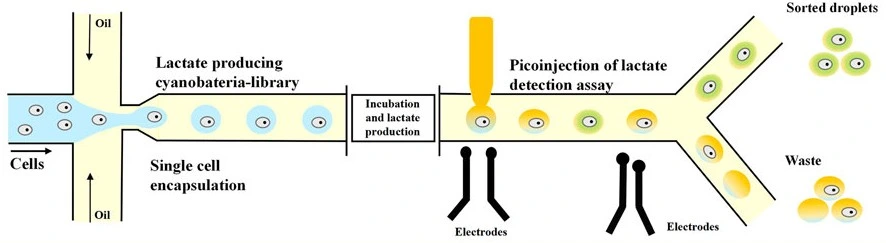
- Hammar, P., et al., Single-cell screening of photosynthetic growth and lactate production by cyanobacteria. Biotechnol Biofuels, 2015. 8: p. 193.
- Ren, C. and A. Lee, History and Current Status of Droplet Microfluidics, in Droplet Microfluidics, C. Ren and A. Lee, Editors. 2020, The Royal Society of Chemistry. p. 0.




Funding and Support
This review was written under the European Union’s Horizon research and innovation program under HORIZON-EIC-2023-PATHFINDEROPEN-01, grant agreement no. 101130747 (Bio-HhOST),
and ANR (Agence Nationale de la Recherche, project number ANR-23-CE10-0018-02) and the SNF (Schweizerischer Nationalfonds) in the framework of the AAPG2023 PRCI (Voxwrite).
This review was written by Celeste Chidiac, PhD.
Published in June 2025.
Contact: Partnership@microfluidic.fr

Check the other Reviews
FAQ - Droplet microfluidics in action: From innovative applications to core challenges
Why choose droplet microfluidics instead of traditional plates and tubes?
Faster responses, better precision, fewer materials wasted – that comes from shrinking processes into tiny droplets, each holding just a fraction of a microliter. Within these small spaces, every step flows smoothly, contained within a single stable unit. Because everything occurs in such confined volumes, detection improves dramatically. Running hundreds at once becomes possible, yet hardly any supplies are used along the way.
How does it compare in performance against micromicrowell arrays or conventional FACS methods?
Timing fluid delivery gets messy with microwell arrays – they work, yet feel awkward. Fast sorting across large cell groups? That belongs to FACS, though it fumbles single-cell details. Tiny droplets change the game: each holds one cell, often capturing what that cell releases, all with little interference. Mix methods rather than choosing one before or after the other, and clarity sharpens while speed stays high.
A specific instance might help – currently, what size makes sense for one cell?
A starting place – genome sequencing of about fifty thousand individual cells, made possible through porous microgels. Then, another angle: monitoring pH and mRNA levels in each rare circulating stromal cell to reveal metabolic traits across patient samples. Each approach stands apart, yet both deliver on true single-cell detail, backed by data at a scale that reflects real biological meaning.
Testing medicines one cell at a time – what drives the move from large-scale methods to droplet-based single-cell tests?
Each drop holds a single cell, offering sharper detail than pooled results ever could. Resistance differs between cells. Inside droplets, substances reach each cell separately: when sensitive ones absorb a lot, they perish; those that resist take in little and remain. Recently developed methods use basic pipettes to build these droplets from small tumor pieces, enabling response measurement exactly where resistance begins. What matters happens cell by cell.
What about using droplets for tissues or organoids – does that fit, or are they just for simpler biological systems?
It is possible now, becoming increasingly workable over time. Using droplets allows the creation of evenly shaped spheroids and organoids while advancing systems that integrate chips capable of managing fluid movement and mechanical pressure. One newer approach, developed recently, generates uniform liver structures from human induced pluripotent stem cells through a technique involving dual-layered droplets and tiny adjustable channels – a method favored when reliable repetition matters more than handcrafted variation.
To what extent does this apply when building DNA structures alongside integrated genomic systems?
Far from basic: combining digital with droplet systems enables automated mixing, consistent nanoliter drops, full-cycle DNA handling – constructing libraries, assembling long fragments, performing barcoded PCR within tiny volumes. Some labs pieced together double-stranded DNA using 160-base segments within droplets; others designed SARS-CoV-2 processes featuring robotic sample prep and chip-based digital droplet detection.
Now here – CRISPR. Could droplets make large-scale editing easier, perhaps even streamline clone selection?
True. Microelectrode arrays move liquid drops through hundreds of channels, zapping them as they go – this links editing with checking in a single setup. For diagnosis, mixing CRISPR tools with microchannels detects signals within thirty minutes using small sample sizes – useful near patients. The system becomes more intricate yet delivers consistent, automated results.
What lies behind the noise in immunotherapy and engineered immune cells – promise or just talk?
It exists. By attaching capture antibodies to the cell surface before enclosing them, researchers can track cytokines right at their source – one cell at a time, without signal overlap. Using paired encapsulation methods allows assessment of interferon-gamma output while preserving the ability to isolate responsive cells later using routine FACS techniques. High transfection rates, near 98%, are achieved when droplets are reshaped or electroporated within microfluidic setups.
What if enzyme development over time correlates with how fast reactions proceed?
Faster droplet tests open doors even with tight funding. One case found improved ACE2 changes right where reactions happen. Another turned an enzyme without metals into one that breaks down certain chemicals nearly a billion times quicker than nature alone – clear proof, not guesswork, guiding choices.
Where do projects usually get stuck?
One challenge keeps appearing: droplets needed for one task often fail elsewhere – tiny ones for cells do not fit larger tissue models. Then there is a mismatch of expectations – the people building devices are rarely the ones using them daily, so small adjustments in routine may require entirely new chips. Materials add another layer – not every option behaves well. Glass allows changes, but costs around 500 USD per unit, too high for many labs. PDMS is inexpensive and lets light through, yet it expands when exposed to oil and shifts during use. Facing these limits sooner avoids long delays later.
Should I build a minimal setup later in the year, or which components would make sense at the start?
Avoid rushing into setup – start by matching the droplet device to what matters most: flow-focusing if uniform drops are critical, or a T-junction where ease counts. For observation, go mild – use basic bright-field imaging combined with just one fluorescent signal. Bring in automation early, choosing either electrodes or valves to replace the trickiest hands-on task. Lock down file names ahead of time. Set clear checkpoints instead of vague goals – track variation in drop size, output rate each hour, and cell health after sorting. Before introducing living cells, test how materials, chemicals, and carrier oils interact across a single-page grid. This steady pattern holds timelines together when experiments drift.
What role does the Microfluidics Innovation Center (MIC) play – could it matter for Horizon Europe bids?
A small company focused on microfluidics, MIC designs droplet-based chips, adds optical or electrical components, builds control systems, automates data collection, and shapes analysis workflows. When part of a Horizon Europe group, an SME with clear expertise sharpens both practical planning and real-world impact; past applications show that MIC involvement is linked to about twice the success rate seen in average submissions. Equipment hurdles slow research less often because prototypes are ready early, freeing collaborators to move straight into publishing results.
What about handling the proposal together while managing integration tasks?
Fine. Our role adjusts TRL levels while structuring tasks tied to single-cell tests, lab-grown tissues in microchips, gene editing processes, or testing biological catalysts – with attention to practical outcomes. In real terms, joint effort shapes the technical chapters. It provides a working model: flow systems, operational controls, and image capture – all recorded clearly so researchers can prioritize discovery rather than setup hurdles.